
Data Structures Information Model
| Issuer: openEHR Specification Program | |
|---|---|
Release: RM Release-1.1.0 |
Status: STABLE |
Revision: [latest_issue] |
Date: [latest_issue_date] |
Keywords: EHR, data structures, reference model, openehr |
|
| © 2003 - 2021 The openEHR Foundation | |
|---|---|
The openEHR Foundation is an independent, non-profit foundation, facilitating the sharing of health records by consumers and clinicians via open specifications, clinical models and open platform implementations. |
|
Licence |
|
Support |
Issues: Problem Reports |
Amendment Record
| Issue | Details | Raiser | Completed |
|---|---|---|---|
RM Release 1.1.0 |
|||
SPECPUB-7: Convert citations to bibtex form. |
T Beale |
||
SPECRM-77. Add |
I McNicoll, |
15 Oct 2019 |
|
SPECPUB-3. Correct UML conversion error in |
S Arikan, |
21 Feb 2019 |
|
RM Release 1.0.4 |
|||
1.7.3 |
SPECRM-83. Improve documentation of |
P Pazos, |
27 Dec 2018 |
RM Release 1.0.3 |
|||
1.7.2 |
SPECRM-42. Clarify definition of |
I McNicoll, |
15 Nov 2015 |
Release 1.0.2 |
|||
1.7.1 |
SPEC-271. Correct minor inconsistencies in |
R Chen |
05 Nov 2008 |
SPEC-257: Correct minor typos and clarify text. |
C Ma, |
||
SPEC-255. Correct minor error in |
A Patterson |
||
SPEC-283. Correct spelling of |
H Frankel |
||
Release 1.0.1 |
|||
1.7 |
SPEC-200. Correct Release 1.0 typographical errors. Minor cosmetic changes to diagrams. Correct return types of |
D Lloyd, |
26 Sep 2006 |
SPEC-207: Change |
S Heard |
||
SPEC-219: Use constants instead of literals to refer to terminology in RM. |
R Chen |
||
SPEC-220: Tighten semantics of |
A Patterson |
||
Release 1.0 |
|||
1.6 |
SPEC-14. Adjust History. Major simplifcation to package; make Events absolute in time. |
S Heard |
16 Dec 2005 |
SPEC-155: Summary data. |
S Heard |
||
SPEC-183. Remove root node from |
G Grieve |
||
SPEC-185. Improved |
S Heard |
||
SPEC-155: Summary data. |
S Heard |
||
SPEC-192: Add display-as-absolute facility to delta Events in History (added explanation only). |
S Heard |
||
SPEC-193: Simplify |
S Heard |
||
SPEC-196: Rename |
S Heard |
||
SPEC-192. Support change, increase and decrease Events in History. |
S Heard |
||
Release 0.96 |
|||
Release 0.95 |
|||
1.5.1 |
SPEC-48. Pre-release review of documents. Fixed |
D Lloyd |
22 Feb 2005 |
1.5 |
SPEC-101. Improve modelling of Structure classes. |
DSTC |
10 Dec 2004 |
SPEC-100. Correct inheritance error in |
T Beale |
||
SPEC-24. Revert meaning to |
S Heard, |
||
SPEC-118. Make package names lower case. |
T Beale |
||
SPEC-123. |
R Chen |
||
SPEC-124. Fix path syntax in data structures IM document. |
T Beale |
||
Release 0.9 |
|||
1.4 |
SPEC-19. Add |
T Beale |
09 Mar 2004 |
SPEC-28. Change name of |
H Frankel |
||
SPEC-89. Remove |
DSTC |
||
SPEC-91. Correct anomalies in use of |
T Beale |
||
SPEC-67. Change |
S Heard |
||
Formally validated using ISE Eiffel 5.4. |
T Beale |
||
1.3.3 |
SPEC-41. Visually differentiate primitive types in openEHR documents. |
D Lloyd |
04 Sep 2003 |
1.3.2 |
SPEC-13 Rename key classes - rename |
D Kalra, |
20 Jun 2003 |
1.3.1 |
Improved heading layout, package naming. Made |
T Beale, |
18 Mar 2003 |
1.3 |
Formally validated using ISE Eiffel 5.2. No changes. |
T Beale |
20 Feb 2003 |
1.2.1 |
Minor corrections to terminology_id invariants. |
Z Tun |
08 Jan 2003 |
1.2 |
Defined packages properly and moved |
T Beale |
18 Dec 2002 |
1.1.1 |
Minor corrections: |
T Beale |
10 Nov 2002 |
1.1 |
Minor adjustments due to change in |
T Beale |
01 Nov 2002 |
1.0 |
Taken from Common RM. |
T Beale |
11 Oct 2002 |
Acknowledgements
The work reported in this paper has been funded in by the following organisations:
-
University College London - Centre for Health Informatics and Multi-professional Education (CHIME);
-
Ocean Informatics;
-
Distributed Systems Technology Centre (DSTC), under the Cooperative Research Centres Program through the Department of the Prime Minister and Cabinet of the Commonwealth Government of Australia.
Thanks to Grahame Grieve of Kestral Computing, Australia for general input and examples relating to History data.
Special thanks to David Ingram, Emeritus Professor of Health Informatics at UCL, who provided a vision and collegial working environment ever since the days of GEHR (1992).
1. Preface
1.1. Purpose
This document describes the common data structures used in openEHR reference model, including lists, tables, trees, and history.
The intended audience includes:
-
Standards bodies producing health informatics standards;
-
Academic groups using openEHR;
-
The open source healthcare community;
-
Solution vendors;
-
Medical informaticians and clinicians interested in health information.
-
Health data managers.
1.3. Status
This specification is in the STABLE state. The development version of this document can be found at https://specifications.openehr.org/releases/RM/latest/data_structures.html.
Known omissions or questions are indicated in the text with a 'to be determined' paragraph, as follows:
TBD: (example To Be Determined paragraph)
1.4. Feedback
Feedback may be provided on the openEHR RM specifications forum.
Issues may be raised on the specifications Problem Report tracker.
To see changes made due to previously reported issues, see the RM component Change Request tracker.
1.5. Conformance
Conformance of a data or software artifact to an openEHR specification is determined by a formal test of that artifact against the relevant openEHR Implementation Technology Specification(s) (ITSs), such as an IDL interface or an XML-schema. Since ITSs are formal derivations from underlying models, ITS conformance indicates model conformance.
2. Background
2.1. Requirements
The requirements for structured data in the EHR and other systems are essentially that low-level data can be expressed in standard structures. The structures which are commonly required are as follows:
-
single values, e.g. weight, height, blood sugar;
-
lists of named/numbered elements, e.g. blood test results;
-
tables of values with named columns and/or named rows, e.g. visual acuity results;
-
trees of values, e.g. biochemistry, microbiology results;
-
histories of values, each of which takes any of the above forms, e.g. a time series of blood pressures, glucose levels, or imaging data.
2.2. Design Principles
The design principle which particularly applies to the data structure models described here is the need to provide explicit specifications for logical structures using the same generic representation, such as hierarchy. The logical structures include tables, lists, trees, and the concept of history.
Regardless of whether such structures are treated as pure presentation or as having semantic significance, there are at various reasons for explicitly including the semantics of logical structures which are represented in a generic way such as hierarchy, including:
-
it is essential for interoperability that a structure such as a logical table, list or linear history be encoded into the generic representation in the same way by all senders and receivers of information, otherwise there is no guarantee that any communicating party’s software processes the structures in the intended fashion;
-
software implementors can develop software which explicitly captures the logical structures as functional interfaces which are used as the only way of building such structures. Such interfaces (assuming they are bug-free) guarantee that all application software always creates correct structures - there is no need to rely on caller software each time making low level calls to create a table or list out of hierarchy elements;
-
the use of a functional interface for such types means that application software at the receiver’s end can always process incoming information in its intended form, enabling correct presentation of data on the screen.
One of the motivations for defining logical data structures explicitly is to remove the ambiguity in recording structure and time in previous EHR specifications and standards, including The Good European Health Record, ISO 13606, HL7 CDA and HL7 FHIR specifications. The approach in these standards to representing time and structure is to use undifferentiated hierarchical structures - there is no specification about how a table or a time series might be represented. Where single values are recorded, an attribute meaning 'time of recording' is usually set; if a time series was required, there is no clear guideline as to how to model it. The standardised approach used here removes all such ambiguity, and improves the quality of data and software.
3. Overview
The rm.data_structures package contains two packages: the item_structure package and
the history package. The first describes generic, path-addressable data structures, while the latter
describes a generic notion of linear history, for recording events in past time. The
data_structures package is illustrated in the UML diagram below.
The data_structures package itself contains a single class, DATA_STRUCTURE, which is the
ancestor of all openEHR data structures. Its only feature is the function as_hierarchy, which is implemented
by each subtype of DATA_STRUCTURE, in order to generate a physical representation of the
structure in ISO 13606 form. The ISO 13606 form is usually less optimal than the openEHR form, but
is compatible with the less semantically rich standard, and is guaranteed (in theory) to be comprehensible
to other systems which support ISO 13606 as an interoperability standard.
3.1. Class Descriptions
3.1.1. DATA_STRUCTURE Class
Class |
DATA_STRUCTURE (abstract) |
|
|---|---|---|
Description |
Abstract parent class of all data structure types. Includes the |
|
Inherit |
||
Functions |
Signature |
Meaning |
1..1 |
as_hierarchy (): |
Hierarchical equivalent of the physical representation of each subtype, compatible with CEN EN 13606 structures. |
4. Item Structure Package
4.1. Overview
The item_structure package classes presented here are a formalisation of the need for generic, archetypable
data structures, and are used by all openEHR reference models.
The subtypes of the ITEM_STRUCTURE class explicitly model the logical data structure types which
typically occur in health record data, and include ITEM_SINGLE (for single values such as a patient
weight), ITEM_LIST (for lists such as parts of an address), ITEM_TREE (for hierarchically structured
data such as a microbiology report) and ITEM_TABLE (for tabular data such as visual acuity or reflex
test results). Each of these classes defines a functional interface, has an optimal physical representation
using the basic types CLUSTER and ELEMENT from the representation package, and can generate
a ISO 13606 compliant hierarchical representation of its data. Any system implementing
these types is guaranteed to create data which represents the logical structures of lists, tables and trees
identically.
Data values are connected to spatial structures via the value attribute of the ELEMENT class of the representation cluster. This class also carries the attribute null_flavour, whose value indicates how to read the contents of the value attribute. Values from the openEHR null flavours vocabulary, including 253|unknown|, 271|no information|, 272|masked|, and 273|not applicable| are used to populate it. Only a small number of generic codes are defined, in order to avoid complex processing for most data instances, for which this simple classification of null is sufficient.
In some circumstances however, additional detail is required in addition to the null flavour code. Examples include reporting and where specific reasons for lack of data have medico-legal ramifications, e.g. 'patient was unconscious', 'patient refused to tell me', 'no reason provided'. For these situations, the optional null_reason field may be used to record a specific reason.
The openEHR class model for spatial structures is illustrated on the right-hand side of Figure 1. It should be noted that these classes (ITEM_LIST etc) are not equivalents of similarly named classes (such as List<T>) in most data structure libraries - they also include per-node name,
archetype_node_id and leaf node value and null flavour, and path capabilities.
4.2. ISO 13606 Encoding Rules
4.2.2. ITEM_LIST
An ITEM_LIST object is encoded in ISO 13606 as a CLUSTER object containing the set of ELEMENTs
from the openEHR list.
4.2.3. ITEM_TABLE
The ITEM_TABLE encoding rules are as follows:
-
Each row is encoded as a Cluster containing a number of
ELEMENTs, each corresponding to the value of a column in that row. -
An empty/void column value for a row is represented by an
ELEMENTcontaining no value, and withnull_flavourset. -
The names of the
ELEMENTin a row are the column names. -
The names of the containing
CLUSTERof each row is the stringified number of the row in the overall table.
4.3. Class Descriptions
4.3.1. ITEM_STRUCTURE Class
Class |
ITEM_STRUCTURE (abstract) |
|
|---|---|---|
Description |
Abstract parent class of all spatial data types. |
|
Inherit |
||
4.3.2. ITEM_SINGLE Class
Class |
ITEM_SINGLE |
|
|---|---|---|
Description |
Logical single value data structure. Used to represent any data which is logically a single value, such as a person’s height or weight. |
|
Inherit |
||
Attributes |
Signature |
Meaning |
1..1 |
item: |
|
Functions |
Signature |
Meaning |
1..1 |
as_hierarchy (): |
Generate a CEN EN13606-compatible hierarchy consisting of a single |
4.3.3. ITEM_LIST Class
Class |
ITEM_LIST |
|
|---|---|---|
Description |
Logical list data structure, where each item has a value and can be referred to by a name and a positional index in the list. The list may be empty.
Not to be used for time-based lists, which should be represented with the proper temporal class, i.e. |
|
Inherit |
||
Attributes |
Signature |
Meaning |
0..1 |
Physical representation of the list. |
|
Functions |
Signature |
Meaning |
1..1 |
item_count (): |
Count of all items. |
0..1 |
Retrieve the names of all items. |
|
1..1 |
Retrieve the item with name ‘a_name’. |
|
1..1 |
Retrieve the i-th item with name. |
|
1..1 |
as_hierarchy (): |
Generate a CEN EN13606-compatible hierarchy consisting of a single |
Invariants |
Valid_structure: ` items.forall (i:ITEM | i.type = "ELEMENT")` |
|
4.3.4. ITEM_TABLE Class
Class |
ITEM_TABLE |
|
|---|---|---|
Description |
Logical relational database style table data structure, in which columns are named and ordered with respect to each other. Implemented using Cluster-per-row encoding. Each row Cluster must have an identical number of Elements, each of which in turn must have identical names and value types in the corresponding positions in each row. Some columns may be designated key' columns, containing key data for each row, in the manner of relational tables. This allows row-naming, where each row represents a body site, a blood antigen etc. All values in a column have the same data type. Used for representing any data which is logically a table of values, such as blood pressure, most protocols, many blood tests etc. Misuse: Not to be used for time-based data, which should be represented with the temporal class |
|
Inherit |
||
Attributes |
Signature |
Meaning |
0..1 |
Physical representation of the table as a list of |
|
Functions |
Signature |
Meaning |
1..1 |
row_count (): |
Number of rows in the table. |
1..1 |
column_count (): |
Return number of columns in the table. |
0..1 |
Return set of row names. |
|
0..1 |
Return set of column names. |
|
1..1 |
Return i-th row. |
|
1..1 |
Return |
|
1..1 |
Return |
|
1..1 |
Return row with name = |
|
1..1 |
Return |
|
1..1 |
Return rows with particular keys. |
|
1..1 |
element_at_cell_ij ( |
Return cell at a particular location. |
1..1 |
as_hierarchy (): |
Generate a CEN EN13606-compatible hierarchy consisting of a single |
Invariants |
Valid_structure: |
|
4.3.5. ITEM_TREE Class
Class |
ITEM_TREE |
|
|---|---|---|
Description |
Logical tree data structure. The tree may be empty. Used for representing data which are logically a tree such as audiology results, microbiology results, biochemistry results. |
|
Inherit |
||
Attributes |
Signature |
Meaning |
0..1 |
The items comprising the |
|
Functions |
Signature |
Meaning |
1..1 |
True if path a_path' is a valid leaf path. |
|
1..1 |
Return the leaf element at the path a_path'. |
|
1..1 |
as_hierarchy (): |
Generate a CEN EN13606-compatible hierarchy, which is the same as the tree’s physical representation. |
4.4. Instance Structures
4.4.1. ITEM_SINGLE Instance Structure
The figure below illustrates a ITEM_SINGLE instance, in both physical and logical forms.
ITEM_SINGLE4.4.2. ITEM_LIST Instance Structure
The following figure illustrates a typical ITEM_LIST structure, in this case for a BP protocol.
ITEM_LIST5. Representation Package
5.1. Overview
This package contains classes for a simple hierarchical representation of any data structure, as shown on the lower right side of Figure 1. These classes are compatible with the ISO 13606-1 classes of the same names, and instances can be losslessly generated to and from ISO 13606 instance structures.
5.2. Class Descriptions
5.2.1. ITEM Class
Class |
ITEM (abstract) |
|
|---|---|---|
Description |
The abstract parent of |
|
Inherit |
||
5.2.2. CLUSTER Class
Class |
CLUSTER |
|
|---|---|---|
Description |
The grouping variant of |
|
Inherit |
||
Attributes |
Signature |
Meaning |
1..1 |
Ordered list of items - |
|
5.2.3. ELEMENT Class
Class |
ELEMENT |
|
|---|---|---|
Description |
The leaf variant of |
|
Inherit |
||
Attributes |
Signature |
Meaning |
0..1 |
null_flavour: |
Flavour of null value, e.g. |
0..1 |
value: |
Property representing leaf value object of |
0..1 |
null_reason: |
Optional specific reason for null value; if set, |
Functions |
Signature |
Meaning |
1..1 |
is_null (): |
True if value logically not known, e.g. if indeterminate, not asked etc. |
Invariants |
Inv_is_null_valid: |
|
Inv_null_flavour_indicated: |
||
Inv_null_flavour_valid: |
||
Inv_null_reason_valid: |
||
6. History Package
6.1. Overview
The history package defines classes which formalise the concept of past, linear time, via which historical data of any structural complexity can be recorded. It supports both instantaneous and interval event samples within periodic and aperiodic series.
Regardless of whether the actual data consist of a single sample or many, they are represented in the same way: as a history of events, i.e. as a time series, allowing all software to access data in a uniform way, whether it be a single measurement of weight, a long series of three- or four-dimensional images, or a series of encapsulated multimedia items. The type of data contained in a History structure may be any openEHR data, including text, coded, quantitative, time, or multimedia. In the case of quantitative and date/time data (rm.data_types.quantity package in openEHR Data Types), the data are formally quantitative, but for the vast majority of cases of other types of data (text, coded etc), there is an obvious quantitative interpretation associated with the values. An example is samples whose data values are coded terms such as 'low', 'normal', 'high', 'critical high', representing blood pressure measurements over time.
Instantaneous events are representing using the type POINT_EVENT, for which the value (data attribute) always represents an actual instantaneous value of a datum at a point in time. However, time series data may also be represented using the INTERVAL_EVENT type, enabling values other than 'actual' to be recorded for intervals of time. Events recorded in this form are accordingly tagged with a mathematical function, such as 'actual', 'mean', 'change' and so on, as defined by the openEHR event math function vocabulary.
The model also supports the inclusion of 'summary' data, i.e. a textual or image item which summarises in some way the entire history.
In formal terms, the model defines the constrained generic (otherwise known as 'template' or 'parameterised') types HISTORY<T→ITEM_STRUCTURE>, EVENT<T→ITEM_STRUCTURE>, POINT_EVENT<T>, and INTERVAL_EVENT<T> where the type parameter is constrained to the ITEM_STRUCTURE type, and defines the type of the data recorded in an instance of HISTORY. The effect is that repeated instances of spatially complex data can recur in time, corresponding to the way data are actually measured. An aperiodic series of POINT_EVENT instances would typically be used to represent manual measurements repeated in time. Periodic histories of INTERVAL_EVENT instances would typically be used to represent vital signs monitor output (which is usually delivered in averaged form potentially with additional minimum and maximum values).
As with all other parts of the openEHR reference model, the history package is designed so that archetypes define the domain semantics of HISTORY instances. The history package is shown on the left of Figure 1.
6.1.1. Basic Semantics
The intention of the History model is to represent time-based data for which every sample in the series is a measurement of the same phenomenon (e.g. patient heart rate) and is obtained using the same measurement method (e.g. pulse oximeter). Samples taken in this way can be reliably treated as comparable, i.e. representing changes in the same phenomenon over time, and accordingly can be safely used for time-based computation, such as graphing, statistical analysis and so on. A History can contain any mixture of POINT_EVENT and INTERVAL_EVENT instances. Clearly it is impossible for the model to guarantee completely correct usage on its own, however there two major safeguards.
Firstly, the use of generic types forces the type of the data in each Event to be the same. A History of type HISTORY<ITEM_LIST> therefore constrains the type of the data at each Event (EVENT.item) to be of type ITEM_LIST and nothing else.
Secondly, the use of archetyping (typically within openEHR Observation archetypes) ensures the actual structure of the ITEM_STRUCTURE subtype is defined in the same way for every sample - e.g. a two-item list representing systolic and diastolic blood pressure.
6.1.2. Timing
An instance of the HISTORY class contains the origin: DV_DATE_TIME attribute, indicating what is considered the '0-point' of the time series, and a series of instances of the EVENT subtype, each containing a time: DV_DATE_TIME attribute representing the absolute time of the event. The relative offset
of any Event is computed as the difference between the EVENT.time and HISTORY.origin by the EVENT.offset function. For Interval events (i.e. instances of INTERVAL_EVENT), the time attribute always refers to the end time of the event, since this is the time at which the data (e.g. average) are true.
The origin time of a History does not have to be the time of the first sample - it might be the time of an event such as child-birth with respect to which the samples are recorded, e.g. Apgar scores (Apgar is a 0-10 score indicating the health of a newborn based on breathing, heartrate, colour, muscletone and reflexes) at 1 and 3 minute offsets.
Periodicity and aperiodicity are expressed via the is_periodic and period attributes. For a periodic time-series, period is set to the period duration value and is_periodic returns True. The total duration of the History is given by the HISTORY.duration function. The following figure illustrates a number of variations in History periodicity and Event type.
6.1.3. Point Events
The simplest kind of Event in a History is a 'point' event, expressed by instances of the class POINT_EVENT, representing an instantaneous value. A HISTORY instance may be composed solely of Point events, as would be the case with a number of blood pressure values measured over time as the patient changes position. An Apgar result is a typical example of aperiodic point data, typically consisting of 2 or 3 events, each containing 5 values and a 6th value repersenting the Apgar score for that time point. Point data may also be available from monitoring devices. For fine-granularity (e.g. 1 second) data, the number of samples may be too voluminous for the health record, and more efficient recording in the form of summary Interval events (see below) might be desired. The diagram below illustrates the structure of a HISTORY containing POINT_EVENTs.
HISTORY<T> of POINT_EVENTs6.1.4. Interval Events
Instances of the INTERVAL_EVENT class are used to express values corresponding to an interval in time. The INTERVAL_EVENT.width attribute defines the duration of the interval; and the inherited time value corresponds to the trailing edge of the event.
The semantics of the Interval event type in this model are that the values effectively summarise actual instantaneous values of a datum that have occurred during the period of the event interval. The mathematical meaning of the data value of any particular interval event is given by the math_function attribute. This is coded by the openEHR vocabulary event math function, and takes values such as 145|minimum|, 144|maximum|, 146|mean|, 147|change| and so on. The math function value on a particular event applies to all the data points attached to the event data attribute.
The particular math functions used in each Interval event in a History may vary throughout the History; for example, one 4-hour Interval event might contain data representing average values, while a following event might contain data representing maximum values for the same or a later time point. Such data can be conveniently used for generating sophisticated graphs of the underlying datum over time. The next figure illustrates a History containing 2 pairs of 4-hour blood pressure Interval events, with each pair containing maximum and mean blood pressure value structures for +4h and +8h timepoints (each of which consist of a systolic and diastolic value).
HISTORY<T> of INTERVAL_EVENTsInterval events can overlap other interval or point events within the same History. A situation where this may occur is with measurement of different periods of vital signs, such as 4-hourly blood pressure events, overlapped by a 24-hour event which contains the values over a period of 6 x 4 hour periods. In general a long Interval event can overlap any combination of Point or Interval events, as shown in the following figure.
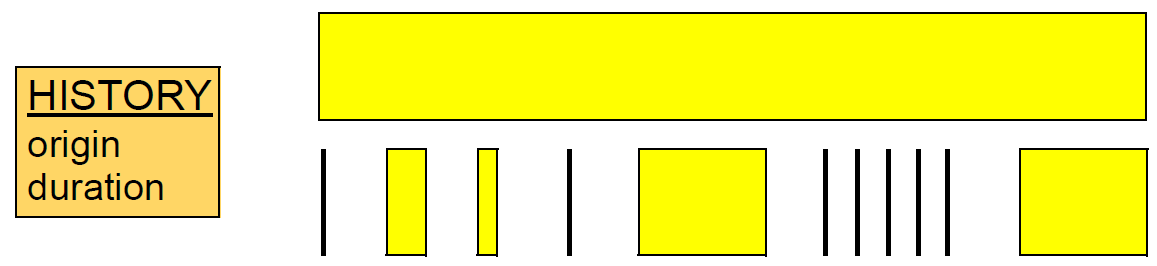
6.1.5. Change Data
One sub-category of interval data that deserves mention is change data. There are three event math function terms used for indicating changes in data values as follows:
-
147|change|: this means that the value recorded is the difference between the value now and the value some time previously. It can be positive or negative; -
522|increase|: the value recorded for the change is positive. The name (i.e.ELEMENT.name) chosen for the item in an archetype carries the semantic of positivity e.g. 'increase of ….; rise of….; ….gain' etc; -
521|decrease|: the value recorded for the change is positive. But the name chosen for the item carries the semantic of negativity e.g. 'decrease of ….; fall of ….; …. loss'.
The following examples show how the data and these math functions are coordinated.
-
weight last week was 76 kg. Wait this week = 74 kg. Possible instances:
| Item Name in Archetype |
Value stored |
Type | Math Function |
|---|---|---|---|
'weight change' |
+ 2kg |
|
|
'weight loss' |
(+)2kg |
|
|
'weight loss' |
True |
|
|
-
weight last week was 80 kg. Weight this week = 83 kg. Possible instances:
| Item Name in Archetype |
Value stored |
Type | Math Function |
|---|---|---|---|
'weight change' |
(+)3kg |
|
|
'weight increase' |
(+)3kg |
|
|
'weight gain' |
True |
|
|
The use of these math function indicators allows the correct representation of change values, no matter how they were captured, in a computable form.
6.1.6. Summary Event Data
A relatively common situation particularly in laboratory testing is the existence of a "summarising" event which accompanies more detailed historical data. Examples where this arises include:
-
a series of exams with a single radiologist report for all of them (the report might include one or more key images);
-
graphical summary of a dynamic challenge test such as Glucose tolerance test;
-
some comment about the pattern of values on a set of observed values in series.
Such data are accommodated within the model via the optional HISTORY.summary attribute, which is
itself a structure, archetypable separately from the structure of the main data. In the first example
above, the summary data might consist of an ITEM_SINGLE object containing a textual report; in the
second, an ITEM_SINGLE object containing a image within a DV_MULTIMEDIA instance.
6.1.7. Efficient Representation of Fine-grained Device Data
A useful practical consequence of the Interval Event is that it allows long periods of stable data to be represented with a single Interval event, while interesting perturbations will be represented with a number of fine-grained Interval or Point Events. In the example in the next figure, Event instances are used represent 4 hours of data consisting to 14,400 x 1 second samples from a blood pressure monitor. The optional INTERVAL_EVENT.sample_count attribute can be used to record the number of original samples summarised in the event. In the illustration, the math_function is shown as 146|mean|; clearly in the first long period, the monitored datum was not absolutely flat. The implication is that the recording software was configured to regard variations in a small band (e.g. 5mm Hg) as insignificant, and only to create new Event objects when the underlying values moved outside the band. Another approach woould have been to create two Interval Event objects for each long period, one giving minimum value, the other maxium value, still based on the principle of generating one such pair for periods when the underlying data remained within specified limits. Regardless of the details, this general approach provides a way to include hours of fine-grained data from devices like vital signs monitors in very little space; the data simply need to be transfomed into equivalent openEHR History form first.
6.1.8. State
A feature particular to a model of recording historical data for scientific and clinical use is the ability to record 'state'. In openEHR, 'state' is understood as time-based information about the whole entity from which the values recorded in the data attribute are recorded. The state attribute thus records data necessary for the correct interpretation of the primary values recorded in the data attribute. An example is where the primary datum is heart rate (values representing rate of the heart beating). Useful state data may include anything else from the owning entity (a human being) that affects the heart rate, such as the level of exertion of the subject (resting, after 3 minutes hard cycling etc). In clinical situations, the state data are often crucial to the safe use of the primary data, since the latter might be normal or abnormal depending on the patient state.
In openEHR there are two ways of recording state. One is via the use of a separate HISTORY structure within the OBSERVATION class (see ehr.composition.content.entry package). The other is via the use of the state attribute of type ITEM_STRUCTURE defined in the class EVENT itself. Experience with openEHR archetypes and systems has shown that the latter method corresponds to the most common clinical need, which is to be able to record the state at the time of the event (the other method allows for the recording of independent state events). A simple example is the recording of 3 glucose levels during a glucose tolerance test. The state information for each event is, respectively (in a typical test):
-
0-minute sample: 'post 8-hour fast;
-
1-hour sample: 'post 75g oral glucose challenge';
-
2-hour sample: 'post 75g oral glucose challenge'.
The History structure for this example is illustrated in the following figure.
HISTORY6.2. Class Descriptions
6.2.1. HISTORY Class
Class |
HISTORY<T> |
|
|---|---|---|
Description |
Root object of a linear history, i.e. time series structure. This is a generic class whose type parameter must be a descendant of For a periodic series of events, period will be set, and the time of each Event in the History must correspond; i.e. the |
|
Inherit |
||
Attributes |
Signature |
Meaning |
1..1 |
origin: |
Time origin of this event history. The first event is not necessarily at the origin point. |
0..1 |
period: |
Period between samples in this segment if periodic. |
0..1 |
duration: |
Duration of the entire History; either corresponds to the duration of all the events, and/or the duration represented by the summary, if it exists. |
0..1 |
summary: |
Optional summary data that aggregates, organizes, reduces and transforms the event series. This may be a text or image that presents a graphical presentation, or some data that assists with the interpretation of the data. |
0..1 |
The events in the series. This attribute is of a generic type whose parameter must be a descendant of |
|
Functions |
Signature |
Meaning |
1..1 |
is_periodic (): |
Indicates whether history is periodic. |
Invariants |
Events_valid: |
|
Periodic_validity: |
||
Period_consistency: |
||
6.2.2. EVENT Class
Class |
EVENT<T> (abstract) |
|
|---|---|---|
Description |
Defines the abstract notion of a single event in a series. This class is generic, allowing types to be generated which are locked to particular spatial types, such as |
|
Inherit |
||
Attributes |
Signature |
Meaning |
1..1 |
time: |
Time of this event. If the width is non-zero, it is the time point of the trailing edge of the event. |
0..1 |
state: |
Optional state data for this event. |
1..1 |
data: |
The data of this event. |
Functions |
Signature |
Meaning |
1..1 |
offset (): |
Offset of this event from origin, computed as time.diff(parent.origin). |
Invariants |
Offset_validity1: |
|
6.2.3. POINT_EVENT Class
Class |
POINT_EVENT<T> |
|
|---|---|---|
Description |
Defines a single point event in a series. |
|
Inherit |
||
6.2.4. INTERVAL_EVENT Class
Class |
INTERVAL_EVENT<T> |
|
|---|---|---|
Description |
Defines a single interval event in a series. |
|
Inherit |
||
Attributes |
Signature |
Meaning |
1..1 |
width: |
Duration of the time interval during which the values recorded under |
0..1 |
sample_count: |
Optional count of original samples to which this event corresponds. |
1..1 |
math_function: |
Mathematical function of the data of this event, e.g. maximum, mean etc. Coded using openEHR vocabulary |
Functions |
Signature |
Meaning |
1..1 |
interval_start_time (): |
Start time of the interval of this event. |
Invariants |
Math_function_validity: |
|
Interval_start_time_valid: |
||
