
Architecture Overview
| Issuer: openEHR Specification Program | |
|---|---|
Release: BASE Release-1.2.0 |
Status: STABLE |
Revision: [latest_issue] |
Date: [latest_issue_date] |
Keywords: EHR, openehr, architecture |
|
| © 2003 - 2021 The openEHR Foundation | |
|---|---|
The openEHR Foundation is an independent, non-profit foundation, facilitating the sharing of health records by consumers and clinicians via open specifications, clinical models and open platform implementations. |
|
Licence |
|
Support |
Issues: Problem Reports |
Amendment Record
| Issue | Details | Raiser, Implementer | Completed |
|---|---|---|---|
BASE Release 1.2.0 |
|||
SPECPUB-7: Convert citations to bibtex form; |
T Beale |
||
SPECRM-88. Improve documentation relating to use of |
P Pazos, |
15 Oct 2019 |
|
BASE Release 1.1.0 |
|||
1.2.2 |
SPECBASE-21. Update Architecture Overview with new LANG component, and improved explanation of ITS. Improve package structure description. Replace some diagrams. |
T Beale |
26 Nov 2018 |
SPECRM-80. Improve description of |
H Frankel, |
||
SPECRM-78: Improve documentation on 'plain text' EHR URIs. |
H Frankel |
22 Nov 2018 |
|
SPECBASE-18. Update text in Common IM to latest openPGP specification. |
P Pazos |
17 May 2018 |
|
1.2.1 |
SPECBASE-16. Update Architecture Overview with current high-level elements. Section 5.3.4: Added text indicating BMM usage and alternatives. Section 11.2.4.2: Added JSON format version of example. |
E Sundvall, S Iancu |
09 Nov 2017 |
SPECBASE-15. Add Foundation Types specification to BASE; |
T Beale |
09 Sep 2017 |
|
BASE Release 1.0.3 |
|||
1.2.0 |
SPECRM-33. Clarify specification of DV_EHR_URI scheme. (solves SPECPR-48, SPECPR-50) |
H Frankel, |
07 Dec 2015 |
SPECRM-27. Relax unique name rule in |
H Frankel, |
||
SPECRM-25. Corrections to Architecture Overview specification: text in section 9.2.2 about |
T Beale |
||
SPECRM-28. Corrections to EHR and Common IM documentation. |
R Erens |
||
Release 1.0.2 |
|||
1.1.1 |
SPEC-249. Paths and locators minor errors in Architecture Overview and Common IM. Typos corrected in sections 9.2.2 and 11.3. |
C Ma, |
13 Nov 2008 |
SPEC-257: Correct minor typos and clarify text. Section 9.2.1 para 1 line 2: with -→ within. |
C Ma, |
||
SPEC-284: Correct inconsistencies in naming of |
A Torrisi |
||
Release 1.0.1 |
|||
1.1 |
SPEC-200: Correct package names in RM diagram. |
D Lloyd |
12 Apr 2007 |
SPEC-130: Correct security details in |
T Beale |
||
SPEC-203: Release 1.0 explanatory text improvements. Improved path explanation. Slight re-ordering of main headings. |
T Beale |
||
Path shortcuts. |
H Frankel |
||
Added configuration management and versioning material from Common IM. |
T Beale |
||
Added section on terminology. |
T Beale |
||
Release 1.0 |
|||
1.0 |
Initial Writing - content taken from Roadmap document. |
T Beale |
29 Jan 2006 |
Acknowledgements
The work reported in this paper has been funded in by the following organisations:
-
University College London - Centre for Health Informatics and Multi-professional Education (CHIME);
-
Ocean Informatics.
Special thanks to David Ingram, Emeritus Professor of Health Informatics at UCL who provided a vision and collegial working environment ever since the days of GEHR (1992).
1. Preface
1.1. Purpose
This document provides an overview of the openEHR architecture in terms of a model overview, key global semantics, deployment and integration architectures, relationship to published standards, and finally the approach to building Implementation Technology Specifications (ITSs). Semantics specific to each information, archetype and service model are described in the relevant specification.
The intended audience includes:
-
Standards bodies producing health informatics standards;
-
Academic groups using openEHR;
-
The open source healthcare community;
-
Solution vendors.
This document is the key technical overview of openEHR, and should be read before all other technical documents.
1.2. Status
This specification is in the STABLE state. The development version of this document can be found at https://specifications.openehr.org/releases/BASE/Release-1.2.0/architecture_overview.html.
Known omissions or questions are indicated in the text with a 'to be determined' paragraph, as follows:
TBD: (example To Be Determined paragraph)
1.3. Feedback
Feedback may be provided on the technical mailing list.
Issues may be raised on the specifications Problem Report tracker.
To see changes made due to previously reported issues, see the BASE component Change Request tracker.
2. Overview
This document provides an overview of the openEHR architecture. It commences with a description of the specification project, followed by an overview of the model structure and packages. Key global semantics including security, archetyping, identification, version and paths are then described. The relationship to published standards is described, and finally, the approach to building implementations is outlined.
2.1. The openEHR Specification Program
The openEHR Specification Program is responsible for developing the specifications of openEHR, from which the openEHR Health Computing Platform is implemented. The outputs of the program consist of a number of components, as shown in the diagram below, with each component (marked in blue) consisting of one or more separate specifications.
The components in the left group contain the abstract specifications (also known as the 'Platform Independent Model' or PIM) and are divided into the following:
-
Foundation (BASE): primitive types, definitions, identifiers, other basic types needed across openEHR;
-
Formalisms (LANG, AM, QUERY): various generic formalisms, including BMM (Basic Meta-Model), (ADL (Archetype Definition Language) and AQL (Archetype Query Language);
-
Content (TERM, RM): the models of primary content of the openEHR platform, including demographics, EHR; supporting openEHR Terminology (along with expressions of various ISO, IETF and other vocabularies (language names, territory names, MIME types, etc) that are not typically published in a directly usable format);
-
Process & CDS (PROC, CDS): clinical process and clinical decision support components, containing the Task Planning and GDL (Guideline Definition Language) specifications;
-
Platform Services: abstract formal APIs defining the interfaces to the openEHR platform.
Some of these specifications (e.g. Antlr grammars) are directly usable in software development. Most of the abstract specifications have concrete expressions in formalisms such as JSON, XML schema, openEHR BMM and REST APIs enabling their direct use in development. These are known as Implementation Technology Specifications (ITSs; also known as 'Platform-Specific Models' or PSMs), and are collected in the ITS component shown on the right. They also constitute openEHR’s interoperability specifications.
As implementation technologies change over time the ITS specifications will be replaced, while the main group of abstract specifications will generally only change in response to real-world requirements other than information technology.
The CNF component at the top defines conformance criteria, and is primarily based on ITS artefacts such as REST APIs, XSDs, but also upon AQL queries, archetypes and some other abstract artefacts. It includes a formal definition of the notional openEHR Platform and how to test it in a standard way. This is used as the basis for openEHR product certification and also for procurement tender specification.
The specifications published by openEHR constitute the primary reference for all openEHR semantics. The presentation style is deliberately intended to be clear and semantically close to the ideas being communicated. Accordingly, the specifications do not follow any particular programming language or idiom, but instead use various formalisms and illustrations appropriate to each topic.
Change control is performed by the openEHR Specifications Editorial Committee (SEC), using a formal process based on Problem Reports (PRs) and Change Reqests (CRs), and a formal release cycle. The details are described in the Specifications Program part of the openEHR website.
The openEHR specification documents and related formal artefacts may be found on the specifications home page. The documents are maintained in Asciidoctor source form, and make significant use of included formal elements, including extracted UML class texts and diagrams, as well as grammar files.
3. Aims of the openEHR Architecture
3.1. Overview
The openEHR architecture embodies over 20 years of research from numerous projects and standards from around the world. It has been designed based on requirements captured over many years, including those developed in the EU FP3 Good European Health Record (GEHR) project (1992-1995).
Because the architecture is highly generic, and particularly due to being archetype-driven, it satisfies many requirements outside the original concept of the 'clinical EHR'. For example, the same reference architecture can be used for veterinary health or even care of public infrastructure or listed buildings. This is due to the fact that the reference model embodies only concepts relating to 'service and administrative events relating to a subject of care'; it is in archetypes and templates that specifics of the kinds of care events and subjects of care are defined. In another dimension, although one of the requirements for the openEHR EHR was a 'patient-centric, longitudinal, shared care EHR', it is not limited to this, and can be used in purely episodic, specialist situations, e.g. as a radiology department record system. Requirements for various flavours of 'health care record' can be categorised according to the two dimensions, scope, and kind of subject, as shown below.
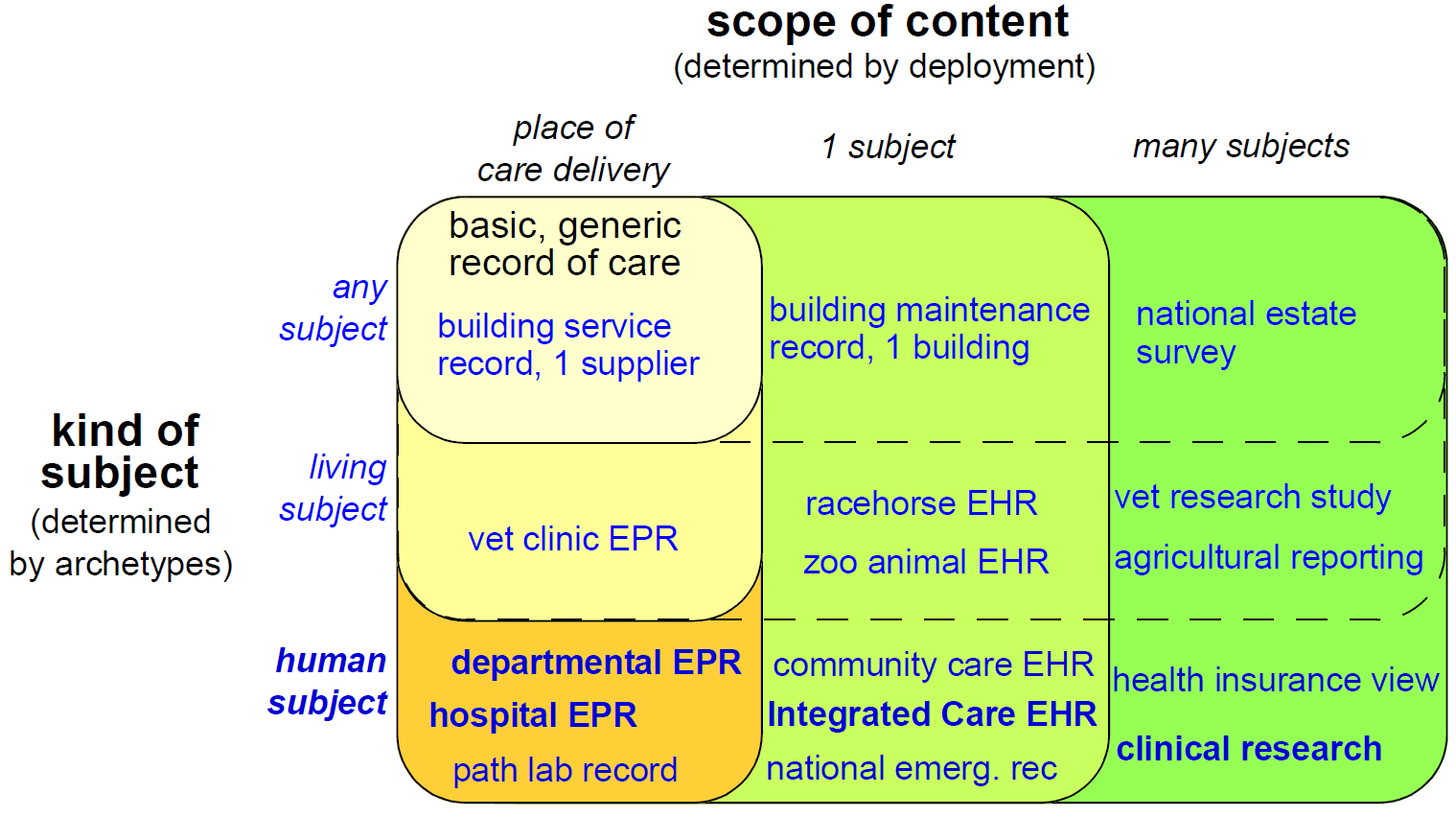
In this figure, each bubble represents a set of requirements, being a superset of all requirements of bubbles contained within it. Requirements for a generic record of care for any kind of subject in a local deployment are represented by the top left bubble. The subsequent addition of requirements corresponding to living subjects and then human subjects is represented by the bubbles down the left side of the diagram. The requirements represented by the largest bubble on the left hand side correspond to 'local health records for human care', such as radiology records, hospital EPRs and so on. Additional sets of requirements represented by wider bubbles going across the diagram correspond to extending the scope of the content of the care record first to a whole subject (resulting in a patient-centric, longitudinal health record) and then to populations or cohorts of subjects, as done in population health and research. From the (human) healthcare point of view, the important requirements groups extend all the way to the bottom row of the diagram.
Going down the diagram, requirements corresponding to increasing specificity of subject of care (from 'any' to 'human') are mostly implemented in openEHR by the use of archetypes. Going across the diagram, the requirements corresponding to increasing scope of record content (from episodic to population) are mainly expressed in different deployments, generally going from standalone to a shared interoperable form. One of the key aspirations for EHRs today is the 'integrated care record' sought by many health authorities today (see ISO 20514 for definition of ICEHR), which provides an informational framework for integrated shared care.
As a result of the approach taken by openEHR, components and applications built to satisfy the requirements of an integrated shared care record can also be deployed as (for example) an episodic radiology record system.
Some of the key requirements developed during the evolution of GEHR to openEHR are listed in the following sections, corresponding to some of the major requirements groups of Figure 2.
3.1.1. Generic Care Record Requirements
The openEHR requirements include the following, corresponding to a basic, generic record of care:
-
prioritisation of the patient / carer interaction (over e.g. research use of the record);
-
suitable for all care settings (primary, acute etc.);
-
medico-legal faithfulness, traceability, audit-trailing;
-
technology & data format independence;
-
highly maintainable and flexible software;
-
support for clinical data structures: lists, tables, time-series, including point and interval events.
3.1.2. Health Care Record (EPR)
The following requirements addressed in openEHR correspond to a local health record, or EPR:
-
support for all aspects of pathology data, including normal ranges, alternative systems of units etc.;
-
supports all natural languages, as well as translations between languages in the record;
-
integrates with any/multiple terminologies.
3.1.3. Shared Care EHR
The following requirements addressed in openEHR correspond to an integrated shared care EHR:
-
support for patient privacy, including anonymous EHRs;
-
facilitate sharing of EHRs via interoperability at data and knowledge levels;
-
compatibility with ISO 13606, Corbamed, and messaging systems;
-
support semi-automated and automated distributed workflows.
3.2. Clinical Aims
From a more specifically clinical care perspective (rather than a record-keeping perspective), the following requirements have been identified during the development of openEHR:
-
The need for a patient-centric, lifelong electronic health record that entails a holistic view of patient needs as opposed to niche problem-solving and decision-support techniques for limited diagnostic purposes;
-
Integration of different views of the patient (GP, emergency and acute care, pathology, radiology, computerised patient-order entry, etc.) with the vast body of available knowledge resources (terminologies, clinical guidelines and computerised libraries);
-
Clinical decision-support to improve patient safety and reduced costs through repeated medical investigations;
-
Access to standards-based computing applications.
The Integrated Care EHR holds great promise: to generalise and make widely available the benefits of computerisation that have been demonstrated individually and in isolated settings. These can be summarised as:
-
Reducing adverse events arising from medication errors such as interactions, duplications or inappropriate treatments and the flow-on costs associated with these;
-
Improving the timely access to critical information and reduced clinician time searching for information;
-
Reducing the incidence of patients being overlooked in the healthcare system due to information not being communicated;
-
Reducing the duplication of investigations and other tests and procedures due to results not being available in the local computing environment;
-
Improved prevention and early detection, based on predictive risk factor analysis, which is possible with quality EHR data;
-
Improved decision making through decision support tools with access to the patient’s whole EHR;
-
Improving access to and computation of evidence based guidelines;
-
Increasing targeted health initiatives known to be effective, based on patient criteria; and
-
Reduced hospitalisations and readmissions.
One comprehensive statement of EHR requirements covering many of the above is the ISO 18308, ISO Technical Report 18308 for which an openEHR profile has been created. The requirements summarised above are described in more detail in the openEHR EHR Information Model specification.
3.3. Deployment Environments
Ultimately any software and information architecture only provides utility when deployed. The architecture of openEHR is designed to support the construction of a number of types of system. One of the most important, the integrated shared care health record is illustrated in the figure below.
In this form, the openEHR services are added to the existing IT infrastructure to provide a shared, secure health record for patients that are seen by any number of health providers in their community context. openEHR-enabled systems can also be used to provide EMR/EPR functionality at provider locations. Overall, a number of important categories of system can be implemented using openEHR including the following:
-
shared-care community or regional health service EHRs;
-
summary EHRs at a national, state, province or similar level;
-
small desktop GP systems;
-
hospital EMRs;
-
consolidated and summary EHRs in federation environments;
-
legacy data purification and validation gateways;
-
web-based secure EHR systems for mobile patients.
Systems containing health records in anonymised or pseudonymised form can also be implemented, since the openEHR architecture defines an EHR in which demographic links (e.g. to national registry, or via national healthcare number) are optional. Where such links are used in the institutional EMR or shared EHR context, they can easily be removed in an anonymisation process.
4. Design Principles
The openEHR approach to modelling information, services and domain knowledge is based on a number of design principles, described below. The application of these principles leads to a separation of the models of the openEHR architecture, and consequently, a high level of componentisation. This leads to better maintainability, extensibility, and flexible deployment.
4.1. Ontological Separation
The most basic kind of distinction in any system of models is ontological, i.e. in the levels of abstraction of description of the real world. All models carry some kind of semantic content, but not all semantics are the same, or even of the same category. For example, some part of the SNOMED CT terminology describes types of bacterial infection, sites in the body, and symptoms. An information model might specify a logical type Quantity. A content model might define the model of information collected in an ante-natal examination by a physician. These types of 'information' are qualitatively different, and need to be developed and maintained separately within the overall model eco-system. The figure below illustrates these distinctions, and indicates what parts are built directly into software and databases.
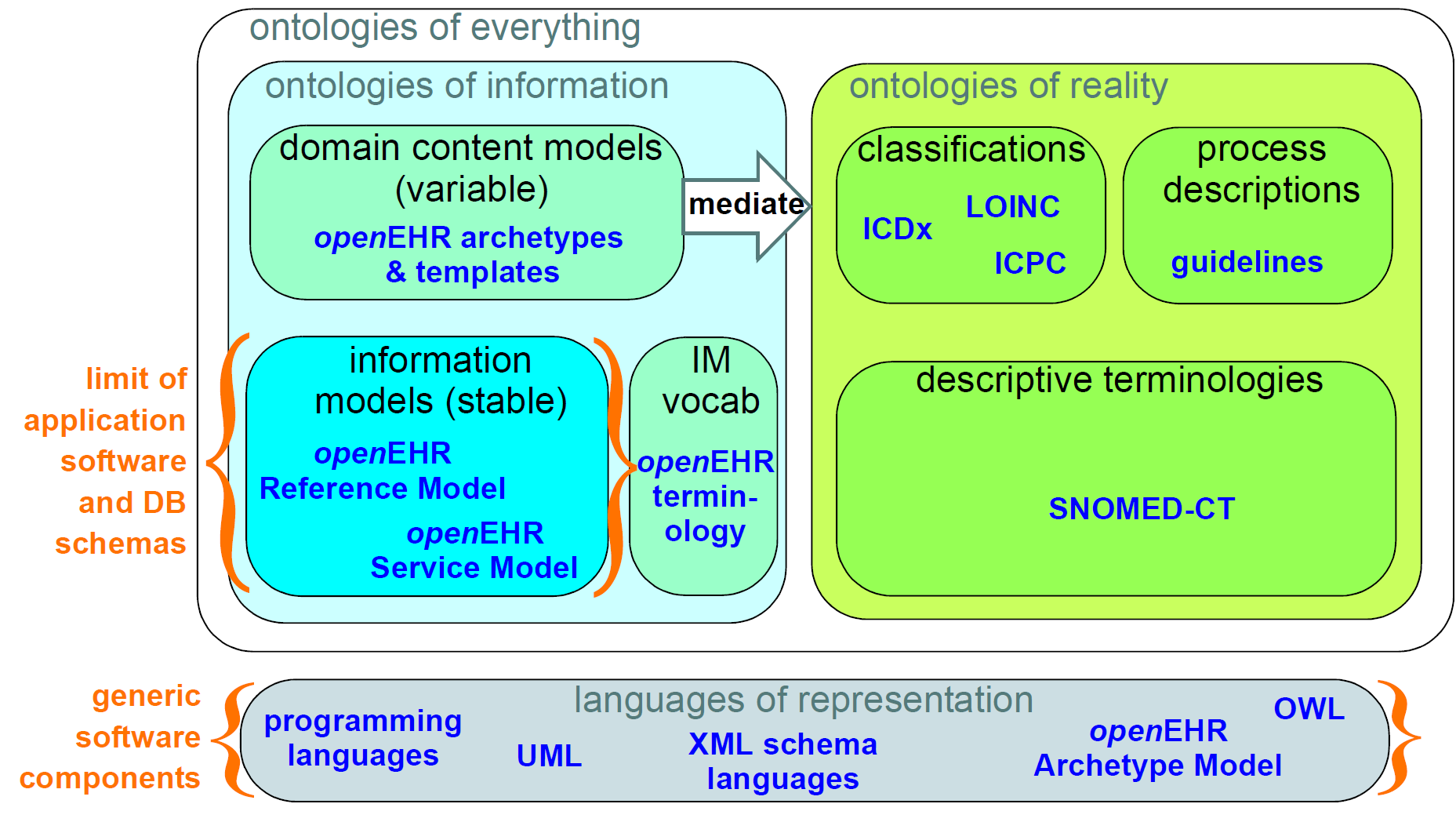
This figure shows a primary separation between 'ontologies of information' i.e. models of information content, and 'ontologies of reality' i.e. descriptions and classifications of real phenomena. These two categories have to be separated because the type of authors, the representation and the purposes are completely different. In health informatics, this separation already exists by and large, due to the development of terminologies and classifications.
A secondary ontological separation within the information side is shown between information models and domain content models. The former category corresponds to semantics that are invariant across the domain (e.g. basic data types like coded terms, data structures like lists, identifiers), while the latter corresponds to variable domain level content descriptions - descriptions of information structures such as 'microbiology result' rather than descriptions of actual phenomena in the real world (such as infection by a microbe). This separation is not generally well understood, and historically, a great deal of domain-level semantics has been hard-wired into the software and databases, leading to relatively unmaintainable systems.
By clearly separating the three categories - information models, domain content models, and terminologies - the openEHR architecture enables each to have a well-defined, limited scope and clear interfaces. This limits the dependence of each on the other, leading to more maintainable and adaptable systems.
4.1.1. Multi-level Modelling and Archetypes
One of the key paradigms on which openEHR is based is known as multi-level modelling, originally described in Beale (2002) as 'two-level modelling'. Under the multi-level approach, there are three levels of models required for a system:
-
reference model (RM): a stable reference information model constitutes the first level of modelling;
-
re-usable content element definitions: formal definitions of clinical content data points and groups, in the form of archetypes;
-
context-specific data set definitions: formal definitions of use-case specific data sets used for forms, documents, messages etc, created by combining required elements of relevant archetypes into openEHR templates.
Only the first level (the Reference Model) is implemented in software, significantly reducing the dependency of deployed systems and data on variable content definitions. The only other parts of the model universe implemented in software are highly stable languages/models of representation (shown at the bottom of Figure 4). As a consequence, systems have the possibility of being far smaller and more maintainable than 'single-level' systems, in which all semantics are expressed in one model (typically a UML class model or DB schema). Archetype-based systems are also inherently self-adapting, since they are built to consume archetypes and templates as they are developed into the future.
Archetypes and templates also act as a well-defined semantic gateway to terminologies, classifications and computerised clinical guidelines. The alternative in the past has been to try to make systems function solely with a combination of hard-wired software and terminology. This approach is flawed, since terminologies don’t contain definitions of domain content (e.g. 'microbiology result'), but rather facts about the real world (e.g. kinds of microbes and the effects of infection in humans); in other words, they are ontological artefacts, whereas archetypes are epistemological artefacts.
The use of archetyping in openEHR engenders new relationships between information and models, as shown in the following figure.
In this figure, 'data' as we know it in normal information systems (shown on the bottom left) conforms in the usual way to an object model (top left). Systems engineered in the 'classic' way (i.e. all domain semantics are encoded somewhere in the software or database) are limited to this kind of architecture. With the use of multi-level modelling, runtime data now conform semantically to archetypes as well as concretely to the reference model. All archetypes are expressed in a generic Archetype Definition Language (ADL), which is the basis of ISO standard 13606-2.
The details of how archetypes and templates work in openEHR are described in the Section 10.
4.1.2. Consequences for Software Engineering
Multi-level modelling significantly changes the dynamics of the systems development process. In the usual IT-intensive process, requirements are gathered via ad hoc discussions with users (typically via the well-known 'use case' methodology), designs and models built from the requirements, implementation proceeds from the design, followed by testing and deployment and ultimately the maintenance part of the lifecycle. This is usually characterised by ongoing high costs of implementation change and/or a widening gap between system capabilities and the requirements at any moment. The approach also suffers from the fact that ad hoc conversations with systems users nearly always fails to reveal underlying content and workflow. Under the multi-level paradigm, the core part of the system is based on the reference and archetype models (includes generic logic for storage, querying, caching etc.), both of which are extremely stable, while domain semantics are mostly delegated to domain specialists who work building archetypes (reusable), templates (local use) and terminology (general use). The process is illustrated in the following figure. Within this process, IT developers concentrate on generic components such as data management and interoperability, while groups of domain experts work outside the software development process, generating definitions that are used by systems at runtime.
Clearly applications cannot always be totally generic (although many data capture and viewing applications are); decision support, administrative, scheduling and many other applications still require custom engineering. However, all such applications can now rely on an archetype- and template-driven computing platform. A key result of this approach is that archetypes now constitute a technology-independent, single-source expression of domain semantics, used to drive database schemas, software logic, GUI screen definitions, message schemas and all other technical expressions of the semantics.
4.2. Separation of Responsibilities
A second design paradigm used in openEHR is that of separation of responsibilities within the computing environment. Complex domains are only tractable if the functionality is first partitioned into broad areas of interest, i.e. into a 'system of systems' (Maier, 2000). This principle has been understood in computer science for a long time under the rubrics 'low coupling', 'encapsulation' and 'componentisation', and was the driver for the explosion of object-oriented languages, libraries and frameworks.
When applied to larger systems, such as that needed to run a hospital or regional health network, the modern form of the paradigm is Services Oriented Architecture (SOA), whereby the components of the system are coarse-grained services. In this approach, each area of functionality is formally modelled and implemented as a self-standing service with a defined interface.
The following diagram illustrates a healthcare services environment containing services at three deployment levels: provider organisation (hospital, clinic, etc); care network (e.g. regional health service, but also non-geographical HMO); and national. These levels may be understood as relating to three perspectives of care (indicated by the larger grey text):
-
healthcare delivery: what happens at a provider enterprise, such as a clinic or hospital;
-
continuity of care: the passage of the patient through multiple clinics and encounters to achieve a care process designed to fulfill a goal;
-
healthcare system: the perspective of a national healthcare system, including public health, planning, quality reporting, etc.
Within each of these deployment levels there are semantic categories corresponding to data, information, process (planning and logistics) and analytics. From left to right, the services are also classified according to what kind of entity they are concerned with: single patient, healthcare professional (HCP), provider enterprise, or knowledge. The diagram is only partially populated, and is not intended to be either complete or normative in any sense.
E-health services at the care network level are emerging, and in many geographies and health organisations, most of the services shown at this level are available only within provider organisations.
Services that openEHR is concerned with specifying (including adaptation of published de jure or other standards) are shown in colour, with other services and applications (e.g. Terminology, patient portal) in grey. As can be seen, the scope of openEHR in terms of services is primarily as follows:
-
patient-centric services at the data and process levels in any deployment level;
-
enterprise-centric services within a care network or provider organisation;
-
knowledge services relating to models of content and process.
Since there are standards available for some aspects of many services, such as terminology, imaging, messages, EHR Extracts, service-based interoperation, and numerous standards for details such as date/time formats and string encoding, the openEHR specifications sometimes act as a mechanism to adapt and integrate existing standards.
4.3. Separation of Viewpoints
The third computing paradigm used in openEHR is a natural consequence of the separation of responsibilities, namely the separation of viewpoints. When responsibilities are divided up among distinct components, it becomes necessary to define a) the information that each processes, and b) how they will communicate. These two aspects of models constitute the two central 'viewpoints' of the ISO RM/ODP model, marked in bold in the following:
| Enterprise |
concerned with the business activities, i.e. purpose, scope and policies of the specified system. |
| Information |
concerned with the semantics of information that needs to be stored and processed in the system. |
| Computational |
concerned with the description of the system as a set of objects that interact at interfaces - enabling system distribution. |
| Engineering |
concerned with the mechanisms supporting system distribution. |
| Technological |
concerned with the detail of the components from which the distributed system is constructed. |
The openEHR specifications accordingly include an information viewpoint - the openEHR Reference Model - and a computational viewpoint - the openEHR Service Model. The Engineering viewpoint corresponds to the openEHR Implementation Technology Specifications (ITS), while the Technological viewpoint corresponds to the technologies and components used in an actual deployment. An important aspect of the division into viewpoints is that there is generally not a 1:1 relationship between model specifications in each viewpoint. For example, there might be a concept of 'health mandate' (see ISO 13940, Continuity of Care concepts) in the enterprise viewpoint. In the information viewpoint, this might have become a model containing many classes. In the computational viewpoint, the information structures defined in the information viewpoint are likely to recur in multiple services, and there may or may not be a 'health mandate' service. The granularity of services defined in the computational viewpoint corresponds most strongly to divisions of function in an enterprise or region, while the granularity of components in the information view points corresponds to the granularity of mental concepts in the problem space, the latter almost always being more fine-grained.
5. openEHR Specification Structure
5.1. Overview
This section provides an overview of the specifications of the openEHR platform components (i.e. the components on the left side of Figure 1). Each component contains one or more specifications, which come in three types:
-
language specifications, expressed in Antlr and/or other formal grammars;
-
information models, expressed in UML;
-
service models (APIs), expressed in UML.
The specifications are published as generated documents consisting of text and various formal elements including:
-
class texts extracted from UML models;
-
diagrams extracted from UML models;
-
grammar files, typically in Antlr4 syntax;
-
EBNF grammars.
The ITS component consists of concrete artefacts of various forms including:
-
API definitions (JSON / Swagger / Apiary etc).
-
XML schemas;
-
JSON schemas;
-
BMM schemas.
5.2. Consolidated Package Structure
From the software engineering point of view, the consolidated structure of UML packages from the formal specifications that contain them is useful, and is illustrated below. The top-level packages include: base, lang, rm, am, proc and sm. All packages defining detailed models appear inside one of these outer packages, which are conceptually defined within the org.openehr namespace (also represented in UML as packages). In some implementation technologies (e.g. Java / Maven), the org.openehr namespace is formally used.
These packages do not include the specifications for various languages (ADL, AQL, ODIN etc), identification systems, or downstream implementation technology specifications (ITSs), which are included in the corresponding component or else in other components.
In the following sections, diagrams provide a visual association of the various specifications to the top-level UML packages, where it exists.
5.3. Base Component (BASE)
The correspondence between the openEHR BASE component specifications and the UML packages is illustrated below. On the right-hand side, the BASE specifications are shown, of which two are based on UML models. These packages fall within the top-level org.openehr.base UML package, as shown on the left side of the figure.
The base package defines identifiers, data types, data structures and various common design patterns that can be re-used ubiquitously in the rm, am and sm packages. The base packages are shown below.
org.openehr.base package|
Note
|
In RM Release 1.0.3 and earlier releases, the contents of the base package resided in the RM support package.
|
The following sub-sections describe the BASE component specifications.
5.3.1. Foundation Types
The Foundation Types specification provides a guide for integrating openEHR models proper into the type systems of implementation technologies. It is specified by the foundations_types package. This contains the special package primitive_types, which describes inbuilt types assumed by openEHR in external type systems. This provides a basis for determining mappings from openEHR to programming languages. For example such as String.is_empty in openEHR might be mapped to String.empty() in a programming environment.
Other foundation types include basic structures (Array<T>, Hash<K,V> etc), time types, and various types enabling functional concepts (principally lambda expressions) to be expressed in the openEHR specifications.
5.3.2. Base Types
The Base Types specification defines generic openEHR types used in other openEHR components. It is comprised of the definitions, identification, terminology and measurement sub-packages. The semantics defined in these packages allow all other models to use identifiers and to have access to knowledge services like terminology and other reference data.
5.3.3. Resource Model
The Resource Model specification defines a generic 'authored resource' class that carries meta-data relating to:
-
authorship;
-
copyright, licences and other related meta-data;
-
languages and translations;
-
annotations.
The class is used via inheritance to provide types in other models with meta-data to enable instances to be managed as resources with appropriate meta-data.
5.4. Languages Component (LANG)
|
Note
|
Until BASE Release 1.1.0, the contents of the LANG component resided in the BASE component. |
The Languages component contains specifications for a number of generic languages used in openEHR, as follows:
-
ODIN: an object data syntax used in openEHR archetypes (in ADL format), in BMM schemas and generally as a data representation where convenient;
-
BMM: the Basic-Meta Model, a formal, human-readable meta-model language in which other models may be expressed for use with tools;
-
EL: Expression Language, a small specification of predicate logic expressions used in other openEHR specifications.
The following diagram shows the relationship of the UML lang package in the global UML structure (left side) to the LANG component specifications defined in terms of UML (right side).
5.4.1. Basic Meta-Model (BMM)
The BMM specification defines a generic meta-model, suitable for formally expressing object-oriented models, including those of openEHR (RM etc). It is roughly an equivalent of UML’s XMI, but fixes various problems with the latter around generic (template) types, while being significantly less complex and fragile. BMM models can be expressed in the ODIN syntax or any other regular object syntax (JSON etc), and conveniently edited by hand. BMM files are used within tools such as the openEHR ADL Workbench and some of the openEHR tooling software.
The BMM is primarily intended to reduce complexity for tools that consume reference model definitions, but is not the only way to implement such tools. Similar tools can be based directly on the openEHR published UML models, as long as typing, template types and qualified attributes are properly handled. Another alternative means of working with models is via software library implementations of the relevant models (openEHR Reference Model etc).
Consequently, understanding or use of BMM specification or models based on it is not necessary in order to implement openEHR systems. However BMM provides a convenient format for model processing, e.g. to auto-generate code stubs in a new language.
5.4.2. Object Data Instance Notation (ODIN)
The ODIN syntax is used to implement faithful machine serialisation and deserialisation of in-memory object graphs, and is a rough equivalent of JSON, YAML and some kinds of XML. It provides more leaf types than any of these, and also supports in-built typing (required to properly represent dynamic binding of polymorphic attributes) and Xpath-like paths.
5.5. Reference Model Component (RM)
The openEHR RM component is illustrated below. All of its specifications are UML model-based.
The figure below illustrates the rm package structure. The packages are in two categories:
-
domain-related:
ehr,demogaphic,ehr_extract,composition,integration; -
generic:
common,data_structures,data_types,support.
The packages in the latter group are generic, and are used by all openEHR models, in all the outer packages. Together, they provide identification, access to knowledge resources, data types and structures, versioning semantics, and support for archetyping. The packages in the first group define the semantics of enterprise level health information types, including the EHR and demographics.
org.openehr.rm packageEach outer package in the above figure corresponds to one openEHR specification document (with the exception of the EHR and Composition packages, which are both described in the EHR Reference Model document), documenting an "information model" (IM). The package structure will normally be replicated in all ITS expressions, e.g. XML schema, programming languages like Java, C# and Eiffel, and interoperability definitions like WSDL, IDL and .Net.
5.5.1. Package Overview
The following sub-sections provide a brief overview of the rm sub-packages.
5.5.1.1. Support Information Model
|
Note
|
this part of the RM has been moved to the BASE component; see above. |
5.5.1.2. Data Types Information Model
A set of clearly defined data types underlies all other models, and provides a number of general and clinically specific types required for all kinds of health information. The following categories of data types are defined in the data types reference model.
| Basic types |
boolean, state variable. |
| Text |
plain text, coded text, paragraphs. |
| Quantities |
any ordered type including ordinal values (used for representing symbolic ordered values such as "+", "++", "+++"), measured quantities with values and units, and so on; includes Date/times - date, time, date-time types, and partial date/time types. |
| Encapsulated data |
multimedia, parsable content. |
| TIME_specification |
types for specifying times in the future, mainly used in medication orders, e.g. '3 times a day before meals'. |
| Uri |
Unique Resource Identifiers. |
5.5.1.3. Data Structures Information Model
In most openEHR information models, generic data structures are used for expressing content whose particular structure will be defined by archetypes. The generic structures are as follows.
| Single |
single items, used to contain any single value, such as a height or weight. |
| List |
linear lists of named items, such as many pathology test results. |
| Table |
tabular data, including unlimited and limited length tables with named and ordered columns, and potentially named rows. |
| Tree |
tree-shaped data, which may be conceptually a list of lists, or other deep structure. |
| History |
time-series structures, where each time-point can be an entire data structure of any complexity, described by one of the above structure types. Point and interval samples are supported. |
5.5.1.4. Common Information Model
Several concepts that recur in higher level packages are defined in the common package. For example, the classes LOCATABLE and ARCHETYPED provide the link between information and archetype models. The classes ATTESTATION and PARTICIPATION are generic domain concepts that provide a standard way of documenting involvement of clinical professionals and other agents with the EHR, including signing.
The change_control package defines a formal model of change management and versioning which applies to any service that needs to be able to supply previous states of its information, in particular the demographic and EHR services. The key semantics of versioning in openEHR are described in the Section 7.3.3.1 section.
5.5.1.5. Security Information Model
The Security Information Model defines the semantics of access control and privacy setting for information in the EHR.
5.5.1.6. EHR Information Model
The EHR IM includes the ehr and composition packages, and defines the containment and context semantics of the key concepts EHR, COMPOSITION, SECTION, and ENTRY. These classes are the major coarse-grained components of the EHR, and correspond directly to the classes of the same names in ISO 13606-1:2005 and fairly closely to the 'levels' of the same names in the HL7 Clinical Document Architecture (CDA) release 2.0.
5.5.1.7. EHR Extract Information Model
The EHR Extract IM defines how an EHR extract is built from COMPOSITIONs, demographic, and
access control information from the EHR. A number of Extract variations are supported, including
"full openEHR", a simplified form for integration with ISO 13606, and an openEHR/openEHR
synchronisation Extract.
5.5.1.8. Integration Information Model
The Integration model defines the class GENERIC_ENTRY, a subtype of ENTRY used to represent freeform
legacy or external data as a tree. This Entry type has its own archetypes, known as "integration
archetypes", which can be used in concert with clinical archetypes as the basis for a tool-based data
integration system. See Section 14 for more details.
5.5.1.9. Demographics Information Model
The demographic model defines generic concepts of PARTY, ROLE and related details such as contact
addresses. The archetype model defines the semantics of constraint on PARTYs, allowing archetypes
for any type of person, organisation, role and role relationship to be described. This approach provides
a flexible way of including the arbitrary demographic attributes allowed in the OMG HDTF PIDS standard.
5.6. Archetype Model Component (AM)
The openEHR AM component is illustrated below.
The openEHR am package contains the models necessary to describe the semantics of archetypes and templates, and their use within openEHR. There are currently two extant major versions of archetype technology in openEHR: 'ADL 1.4', the original version, and 'ADL 2', a more modern version, which is slowly being adopted. Both versions are maintained side by side, to enable implementers to work with the version(s) that suit their needs.
In both versions, the Archetype Model consists of ADL, the Archetype Definition Language (expressed in the form of a syntax specification), and the Archetype Object Model (AOM), a structured model of archetypes.
The package structure of the version 2 form of the AM is shown below.
org.openehr.am packageThe package structure of the version 1.4 form of the AM is shown below.
org.openehr.am packageAnother key specification is the Archetype Identification specification, which defines semantics for archetype identifiers, versioning and life-cycle. The formal specifications may be found on the Archetype Model index page.
5.7. Service Model (SM)
The openEHR service model includes definitions of basic services in the health information environment, centred around the EHR. It is illustrated in the figure below. The set of services actually included is evolving over time.
org.openehr.sm package5.7.1. Definitions Service
The Definitions Service defines the interface to online repositories of archetypes, templates and AQL queries, and can be used both by GUI applications designed for human browsing as well as access by other software services such as the EHR.
5.7.2. EHR Service
The EHR Service defines the coarse-grained interface to electronic health record service. The level of granularity is openEHR Contributions and Compositions, i.e. a version-control / change-set interface.
Part of the model defines the semantics of server-side querying, i.e. queries which cause large amounts of data to be processed, generally returning small aggregated answers, such as averages, or sets of ids of patients matching a particular criterion.
5.7.3. Query Service
The Query Service defines the interface via which AQL queries which may be stored via the Definitions Service, or ad hoc, can be executed.
5.7.4. Terminology Interface
The Terminology Service service provides the means for all other services to access any terminology available in the health information environment, including basic classification vocabularies such as ICDx and ICPC, as well as more advanced ontology-based terminologies. Following the concept of division of responsibilities in a system-of-systems context, the Terminology Service abstracts the different underlying architectures of each terminology, allowing other services in the environment to access terms in a standard way. The Terminology Service is thus the gateway to all ontology- and terminology- based knowledge services in the environment, which along with services for accessing guidelines, drug data and other "reference data" enables inferencing and decision support to be carried out in the environment.
5.8. Global View
The figure below shows all of the openEHR specifications, i.e. object models, languages and APIs, arranged by component. This view abstracts away the components and top-level UML packages, providing a useful aide memoire picture of the totality of openEHR specifications. Dependencies only exist from higher elements to lower elements. The CNF and ITS components are separated since they are semantically derivative from the primary specifications, but are primary artefacts for downstream software engineering use. The other usable software artefact comes in the form of class libraries directly implementing the formal specifications.
6. Design of the openEHR EHR
6.1. The EHR System
The notion of a logical EHR system is central to the openEHR architecture. In openEHR, a system is understood as a distinct logical repository corresponding to an organisational entity that is legally responsible for the management and governance of the healthcare data contained within. This may be a regional health service that serves multiple provider enterprises or a single provider enterprise such as a larger hospital. The 'system' is therefore in general distinct from specific applications and also from provider organisations, even if in some cases it happens to be owned by a single provider. It is also distinct from any underlying virtualisation infrastructure or cloud computing facility, which may house multiple logical EHR systems in a multi-tenant fashion. This is clear by comparing the contractual responsibilities of the infrastructure provider, which are for generic IT service management, to a procurer (e.g. a healthcare data management entity). It is the latter that undertakes legal responsibility for the content, on behalf of one or more healthcare provider organisations.
The technical criterion for identifying an EHR system is that it is the entity that assigns version identifiers within a repository.
6.1.1. System Identity
Within the openEHR architecture, a system_id attribute is recorded both within each patient EHR (EHR class), and also within the audit created with each commit of data to an EHR (AUDIT_DETAILS class). It is also used in feeder system audits that record the origin of imported data (FEEDER_AUDIT_DETAILS class). This identifier identifies the logical EHR system as described above, and may be of any form. Common forms include the reverse domain name and plain and structured string identifiers.
The system identifier is not assumed to be directly processable, but may instead be used as a key, for example in a service maintaining location information.
6.1.2. Information Architecture
In informational terms, a minimal EHR system based on openEHR consists of an EHR repository, an archetype repository, terminology (if available), and demographic/identity information, as shown below.
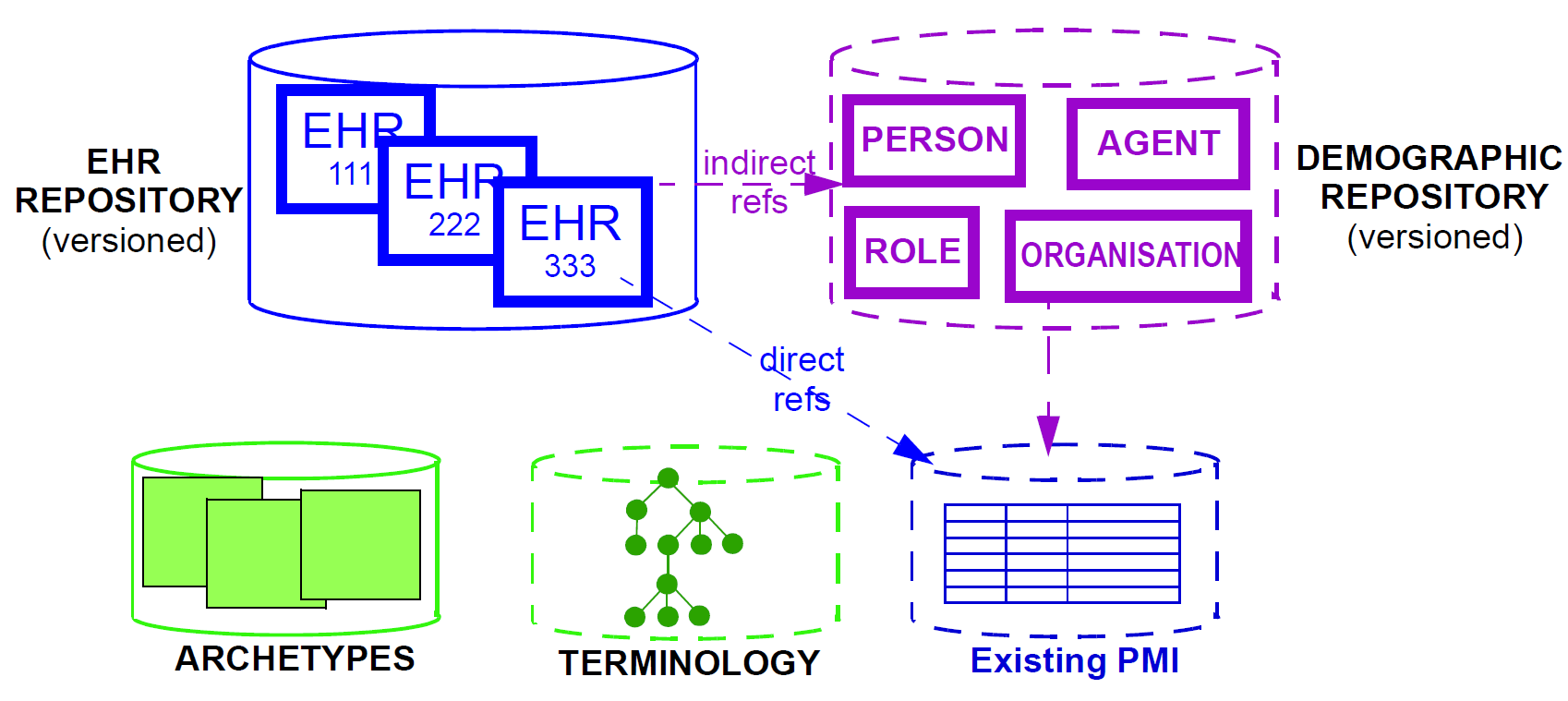
The latter may be in the form of an existing PMI (patient master index) or other directory, or it may be in the form of an openEHR demographic repository. An openEHR demographic repository can act as a front end to an existing PMI or in its own right. Either way it performs two functions: standardisation of demographic information structures and versioning. An openEHR EHR contains references to entities in whichever demographic repository has been configured for use in the environment; the EHR can be configured to include either no demographic or some identifying data. One of the basic principles of openEHR is the complete separation of EHR and demographic information, such that an EHR taken in isolation contains little or no clue as to the identity of the patient it belongs to. The security benefits are described below. In more complete EHR systems, numerous other services (particularly security-related) would normally be deployed, as shown in Figure 7.
6.2. Top-level Information Structures
As has been shown, the openEHR information models define information at varying levels of granularity. Fine-grained structures defined in the Support and Data types are used in the Data Structures and Common models; these are used in turn in the EHR, EHR Extract, Demographic and other 'top-level' models. These latter models define the 'top-level structures' of openEHR, i.e. content structures that can sensibly stand alone, and may be considered the equivalent of separate documents in a document-oriented system. In openEHR information systems, it is generally the top-level structures that are of direct interest to users. The major top-level structures include the following:
| Composition |
the committal unit of the EHR (see type |
| EHR Access |
the EHR-wide access control object (see type |
| EHR Status |
the status summary of the EHR (see type |
| Folder hierarchy |
act as directory structures in EHR, Demographic services (see type FOLDER in Common IM); |
| Party |
various subtypes including |
| EHR Extract |
the transmission unit between EHR systems, containing a serialisation of EHR, demographic and other content (see type |
All persistent openEHR EHR, demographic and related content is found within top-level information structures. Most of these are visible in the following figures.
6.3. The EHR
The openEHR EHR is structured according to a relatively simple model. A central EHR object identified by an EHR id specifies references to a number of types of structured, versioned information, plus a list of Contribution objects that act as audits for changes made to the EHR. The high-level structure of the openEHR EHR is shown below.
In this figure, the parts of the EHR are as follows:
-
EHR: the root object, identified by a globally unique EHR identifier;
-
EHR_access (versioned): an object containing access control settings for the record;
-
EHR_status (versioned): an object containing various status and control information, optionally including the identifier of the subject (i.e. patient) currently associated with the record;
-
Directory (versioned): an optional hierarchical structure of Folders that can be used to logically organise Compositions;
-
Folders (versioned): additional optional hierarchical folder structures that can be used to logically organise Compositions;
-
Compositions (versioned): the containers of all clinical and administrative content of the record;
-
Contributions: the change-set records for every change made to the health record; each Contribution references a set of one or more Versions of any of the versioned items in the record that were committed or attested together by a user to an EHR system.
The logical structure of a typical Composition is shown in more detail in the next figure. This shows various hierarchical levels from Composition to the data types are shown in a typical arrangement. The 21 data types provide for all types of data needed for clinical and administrative recording.
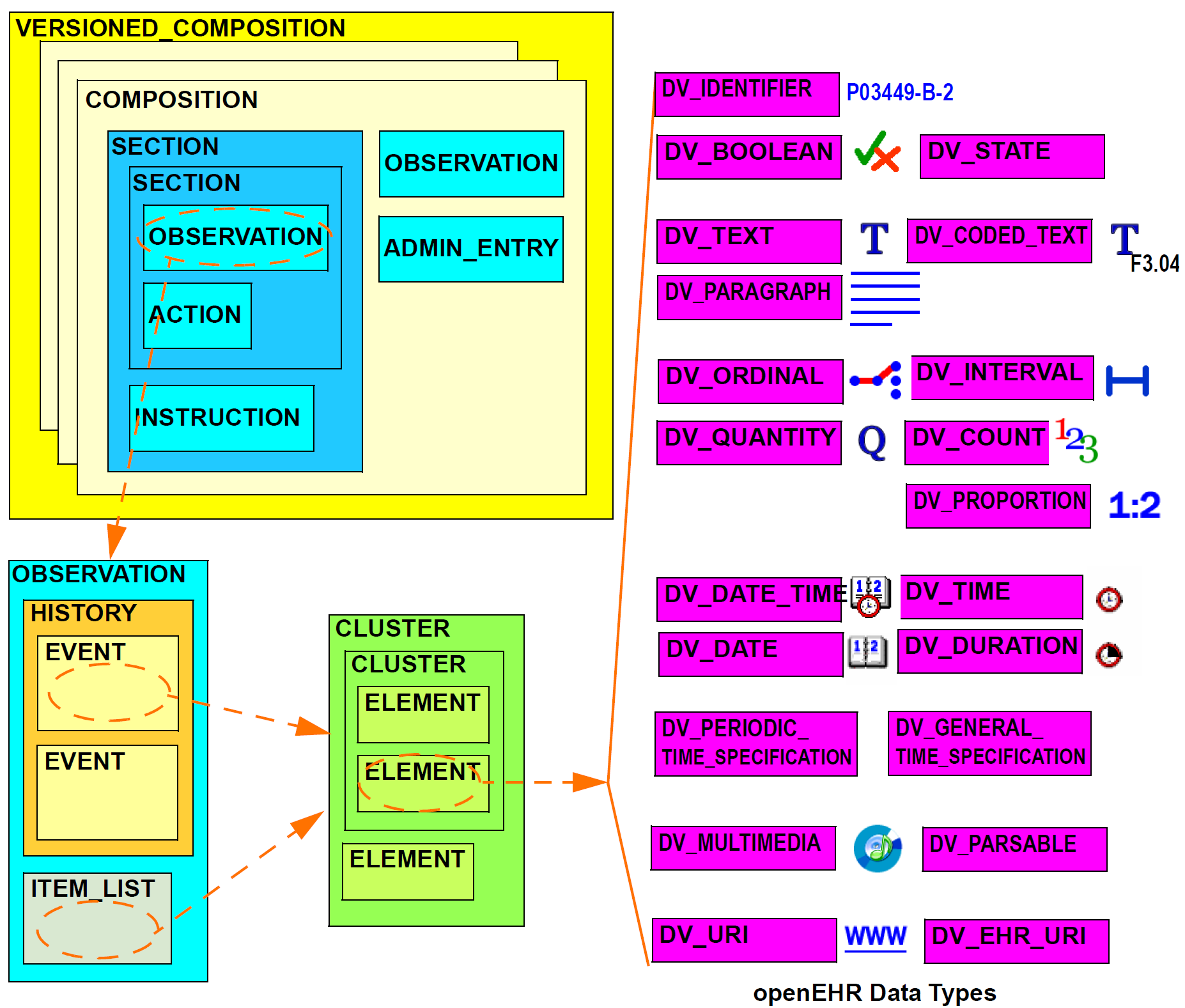
6.4. Entries and Clinical Statements
6.4.1. Entry Subtypes
All clinical information created in the openEHR EHR is ultimately expressed in 'Entries'. An Entry is logically a single clinical statement, and may be a single short narrative phrase, but may also contain a significant amount of data, e.g. an entire microbiology result, a psychiatric examination note, a complex medication order. In terms of actual content, the Entry classes are the most important in the openEHR EHR Information Model, since they define the semantics of all the 'hard' information in the record. They are intended to be archetyped, and in fact, archetypes for Entries and sub-parts of Entries make up the vast majority of archetypes defined for the EHR.
The openEHR ENTRY classes are shown below. There are five concrete subtypes: ADMIN_ENTRY, OBSERVATION, EVALUATION, INSTRUCTION and ACTION, of which the latter four are kinds of CARE_ENTRY.
The choice of these types is based on the clinical problem-solving process, described in Beale & Heard (2007) shown below.
This figure shows the cycle of information creation due to an iterative, problem solving process typical not just of clinical medicine but of science in general. The 'system' as a whole is considered to be made up of two parts: the 'patient system' and the 'clinical investigator system'. The latter consists of health carers, and may include the patient (at points in time when the patient performs observational or therapeutic activities), and is responsible for understanding the state of the patient system and delivering care to it. A problem is solved by making observations, forming opinions (hypotheses), and prescribing actions (instructions) for next steps, which may be further investigation, or may be interventions designed to resolve the problem, and finally, executing the instructions (actions).
This process model is a synthesis of Lawrence Weed’s 'problem-oriented' method of EHR recording, and later related efforts, including the model of Rector Nowlan & Kay (1991), and the 'hypothetico-deductive' model of reasoning (see e.g. Elstein Shulman & Sprafka (1978)). However hypothesis-making and testing is not the only successful process used by clinical professionals - evidence shows that many (particularly those older and more experienced) rely on pattern recognition and direct retrieval of plans used previously with similar patients or prototype models. The investigator process model used in openEHR is compatible with both cognitive approaches, since it does not say how opinions are formed, nor imply any specific number or size of iterations to bring the process to a conclusion, nor even require all steps to be present while iterating (e.g. GPs often prescribe without making a firm diagnosis). Consequently, the openEHR Entry model does not impose a process model, it only provides the possible types of information that might occur.
6.4.1.1. Ontology of Entry Types
In the clinical world practitioners do not think in terms of only five kinds of data corresponding to the subtypes of Entry described above. There are many subtypes of each of these types, of which some are shown in the figure below, reproduced from Beale & Heard (2007).
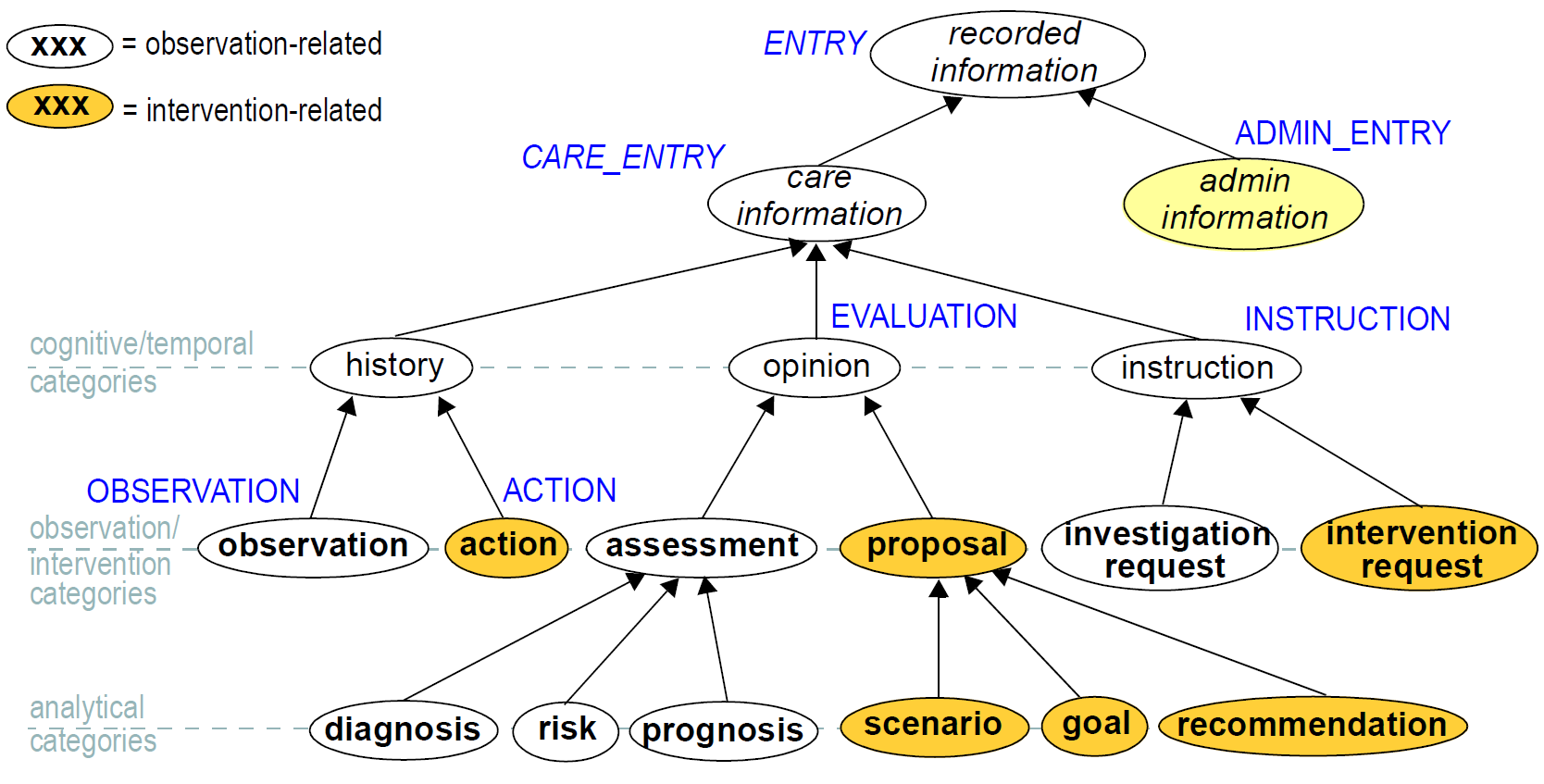
The key top-level categories are 'care information' and 'administrative information'. The former encompasses all statements that might be recorded at any point during the care process, and consists of the major sub-categories on which the Entry model is based, namely 'observation', 'opinion', 'instruction', and 'action' (a kind of observation) which themselves correspond to the past, present and future in time. The administrative information category covers information which is not generated by the care process proper, but relates to organising it, such as appointments and admissions. This information is not about care, but about the logistics of care delivery. Regardless of the diversity, each of the leaf-level categories shown in this figure is ultimately a sub-category of one of the types from the process model, and hence, of the subtypes of the openEHR Entry model.
Correct representation of the categories from the ontology is enabled by using archetypes designed to express the information of interest (say a risk assessment) in terms of a particular Entry subtype (in this case, Evaluation). In a system where Entries are thus modelled, there will be no danger of incorrectly identifying the various kinds of Entries, as long as the Entry subtype, time, and certainty/negation are taken into account. Note that even if the ontology shown in Figure 24 is not correct (undoubtedly it is not), archetypes will be constructed to account for each improved idea of what such categories should really be.
6.4.1.2. Clinical Statement Status and Negation
A well-known problem in clinical information recording is the assignment of 'status' to recorded items. Kinds of status include variants like "actual value of P" (P stands for some phenomenon), "family history of P", "risk of P", "fear of P", as well as negation of any of these, i.e. "not/no P", "no history of P" etc. A proper analysis of these so called statuses shows that they are not "statuses" at all, but different categories of information as per the ontology of Figure 24. In general, negations are handled by using "exclusion" archetypes for the appropriate Entry type. For example, "no allergies" can be modelled using an Evaluation archetype that describes which allergies are excluded for this patient. Another set of statement types that can be confused in systems that do not properly model information categories concern interventions, e.g. "hip replacement (5 years ago)", "hip replacement (recommended)", "hip replacement (ordered for next Tuesday 10 am)".
All of these statement types map directly to one of the openEHR Entry types in an unambiguous fashion, ensuring that querying of the EHR does not match incorrect data, such as a statement about fear or risk, when the query was for an observation of the phenomenon in question.
Further details on the openEHR model clinical information are given in the EHR IM document, Entry Section.
6.5. Managing Interventions
A key part of the investigation process shown in Figure 23, and indeed healthcare in general, is
intervention. Specifying and managing interventions (whether the simplest prescriptions or complex
surgery and therapy) is a hard problem for information systems because it is in 'future time' (meaning
that intervention activities have to be expressed using branching/looping time specifications, not
the simple linear time of observations), unexpected events can change things (e.g. patient reaction to
drugs), and the status of a given intervention can be hard to track, particularly in distributed systems.
However, from the health professional’s point of view, almost nothing is more basic than wanting to
find out: what medications is this patient on, since when, and what is the progress?
The openEHR approach to these challenges is to use the Entry type INSTRUCTION, its subpart
ACTIVITY to specify interventions in the future, and the Entry subtype ACTION to record what has
actually happened. A number of important features are provided in this model, including:
-
a single, flexible way of modelling all interventions, whether they be single drug medication orders or complex hospital-based therapies;
-
a way of knowing the state of any intervention, in terms of the states in a standard state machine, shown below; this allows a patient’s EHR to be queried in a standard way so as to return "all active medications", "all suspended interventions" etc.;
-
a way of mapping particular care process flow steps to the standard state machine states, enabling health professionals to define and view interventions in terms they understand;
-
support for automated workflow, without requiring it.
Coupled with the comprehensive versioning capabilities of openEHR, the Instruction/Action design allows clinical users of the record to define and manage interventions for the patient in a distributed environment.
6.6. Time in the EHR
Time is well-known as a challenging modelling problem in health information. In openEHR, times that are a by-product of the investigation process (e.g. time of sampling or collection; time of measurement, time of a healthcare business event, time of data committal) described above are concretely modelled, while other times specific to particular content (e.g. date of onset, date of resolution) are modelled using archetyping of generic data attributes. The following figure shows a typical relationship of times with respect to the observation process, and the corresponding attributes within the openEHR reference model. Note that under different scenarios, such as GP consultation, radiology reporting and others, the temporal relationships may be quite different than those shown in the figure. Time is described in detail in the EHR Information Model.
6.7. Language
In some situations, there may be more than one language used in the EHR. This may be due to patients being treated across borders (common among the Scandinavian countries, between Brazil and northern neighbours), or due to patients being treated while travelling, or due to multiple languages simply being used in the home environment.
Language is handled as follows in the openEHR EHR. The default language for the whole EHR is determined from the operating system locale. It may be included in the EHR_status object if desired. Language is then mandatorily indicated in two places in the EHR data, namely in Compositions and Entries (i.e. Observations, etc), in a language attribute. This allows both Compositions of different languages in the EHR, and Entries of different languages in the same Composition. Additionally, within Entries, text and coded text items may optionally have language recorded if it is different from the language of the enclosing Entry, or where these types are used within other non-Entry structures that don’t indicate language.
The use of these features is mostly likely to occur due to translation, although in some cases a truly multi-lingual environment might exist within the clinical encounter context. In the former case, some parts of an EHR, e.g. particular Compositions will be translated before or after a clinical encounter to as to make the information available in the primary language of the EHR. The act of translation (like any other interaction with the EHR) will cause changes to the record, in the form of new Versions. New translations can conveniently be recorded as branch versions, attached to the version of which they are a translation. This is not mandatory, but provides a convenient way to store translations so that they don’t appear to replace the original content.
7. Security and Confidentiality
7.1. Requirements
7.1.1. Privacy, Confidentiality and Consent
Privacy (the right to limit who sees the personal data) and confidentiality (the obligation of others to respect the privacy of disclosed data) are primary concerns of many consumers with respect to e- Health systems. A widely accepted principle is that information provided (either directly or due to observation or testing of specimens etc.) in confidence by a patient to health professionals during an episode of care should only be passed on or otherwise become available to other parties if the patient agrees; put more simply: data sharing must be controlled by patient consent. A more complex subrequirement for some patients is allowing differential access to parts of their health record, for example, relatively open access rights to most of the health record, but limited access to sexual or mental health items. The interrelatedness of health information can make this difficult. For example the medication list will often give away sensitive conditions even if the diagnosis is hidden, yet is needed for any safe treatment, and many health professionals would see the unavailability of current medications (and allergies) information as highly problematic for giving even basic care.
7.1.2. Requirements of Healthcare Providers
On the other hand, clinical professionals delivering care want fast access to relevant data, and to be sure that what they see on the screen is a faithful representation of what has been said about the patient. Emergency access to health records is sometimes needed by carers otherwise unrelated to the normal care of a patient; such accesses can only be consented to in a general way, since the specific providers involved will not usually be known.
Researchers in healthcare generally want access to the data of large numbers of patients in order to evaluate current care and improve it (clinical knowledge discovery), and for educational purposes. Both of these latter needs are also ultimately patient and societal priorities. Providing effective care and supporting ongoing medical research therefore have to function in a system that implements the concept of patient consent.
7.1.3. Specifying Access Control
In theory, it should be easy for the patient or some clinical professional to specify who can see the patient record. In some cases it can be done by direct identification, e.g. the patient might nominate their long-term GP by provider id. Some exclusions could potentially be made this way as well, for example a previous doctor with whom the patient has a problematic personal relationship. However it soon becomes difficult to identify provider parties individually when the patient moves into parts of the healthcare system where there are many staff, and/or where there is no previously established relationship. The advent of e-prescribing and e-pharmacy will bring even larger numbers of health and allied health workers into the e-Health matrix, making the problem of individual identification of who should see the patient’s data infeasible. Further, there is a large and growing category of "very mobile" people (the military, entertainers, NGO workers, international business and tourism professionals, athletes….) who cannot predict even in which country they may require care. As a consequence, the need for some access control to be specified in terms of categories or role types appears inescapable.
7.1.4. The Problem of Roles
One of the difficult challenges to implementing access control to health information is that of defining "roles", i.e. the status of users of the record at the time when right of access is being determined. In principle, roles ought to be knowable in advance. For example, the labels "nurse", "GP" and "psychiatrist" can be relatively easily assigned to individuals. However, the kinds of labels that are of more importance are those that differentiate among (for example) personal carers (e.g. primary GP), other care delivery staff (e.g. nurses, aged carers) and support staff (e.g. pathologists, radiographers). In a patient care delivery-oriented view of the world, the professional level of a health care professional is probably less important than his or her relationship to the current care process for the patient.
It will not always be clear which individuals fall into any of these categories at any time, or how such terms are even defined in different sites and jurisdictions. Realistically, the evaluation of a role category such as "care deliverer" into particular identities such as those of nurses on the ward on a particular day must be done in each care delivery environment, not in the EHR. Access decisions for information in the EHR therefore will have some dependence on provider site knowledge of which staff are actively involved in the care process of a given patient.
Role-based access control is further complicated by the common fact of temporary replacements due to illness or holiday and role changes due to staff shortages. Further, if a physician employing a medical secretary requires her to access and update sensitive parts (relating to his own treatment of the patient) of the record, access at the highest level is effectively given to someone not medically trained or related directly to the patient’s care, even if only for 10 minutes. Any role-based system therefore has to take into account the messy reality of clinical care in the real world rather than being based solely on theoretical principles.
7.1.5. Usability
Usability of security and privacy mechanisms is a key requirement of a health record architecture. Some very elegant solutions to fine-grained access control designed by security experts would be simply unusable in practice because they would take too long for patients and doctors to learn, or are too time-consuming to actually use on the screen; they could also be too complex to safely implement in software.
The following sections describe support in openEHR for the main security and privacy requirements of EHRs.
7.2. Threats to Security and Privacy
Any model of how security and privacy are supported in the health record must be based on some notion of assumed threats. Without going into great detail, security threats assumed by openEHR include the following (here "inappropriate" means anything that is not or would not be consented to by the patient):
-
human error in patient identification, leading to incorrect association of health data of one patient with another. Mis-identification of patients can cause personal data for one patient to go into the record of another patient (leading to privacy violations and possibly clinical errors), or into a new record rather than the existing one for the same patient (leading to two or more clinically incomplete records);
-
inappropriate access by health professionals or others in the physical care delivery environment (including e.g. any worker in a hospital) not involved in the current care of the patient;
-
inappropriate access by other persons known to the patient, e.g. a by family member;
-
inappropriate access of health data by corporate or other organisations e.g. for purposes of insurance discrimination;
-
malicious theft of or access to health data (e.g. of a celebrity or politician) for profit or other personal motives;
-
generic threats to data integrity and availability, such as viruses, worms, denial of service attacks etc.;
-
failures in software (due to bugs, incorrect configuration, interoperability failures etc.) causing corruption to data, or incorrect display or computation, resulting in clinical errors.
A key principle with respect to the design of mechanisms supporting security, confidentiality and integrity has to be kept in mind: the likelihood of any given mode of targeted inappropriate access is proportional to the perceived value of the information and inversely proportional to the cost of access. To paraphrase Ross Anderson’s BMA paper (Anderson & Anderson, 1996) on health data security, for a given access, the perpretrator will try to find the simplest, cheapest and quickest method, which is more likely to be bribery or burglary than James Bond-inspired technology. openEHR makes use of this principle by providing some relatively simple mechanisms that are cheap to implement but can make misuse quite difficult, without compromising availability.
7.3. Solutions Provided by openEHR
7.3.1. Overview
Many of the concrete mechanisms relating to security and privacy are found in system deployments rather than in models such as openEHR, particularly the implementation of authentication, access control, and encryption. The openEHR specifications and core component implementations do not explicitly define many concrete mechanisms since there is great variability in the requirements of different sites - secure LAN deployments many require minimal security inside, whereas web-accessible health record servers are likely to have quite different requirements. What openEHR does is to support some of the key requirements in a flexible enough way that deployments with substantially different requirements and configurations can nevertheless implement the basic principles in a standard way.
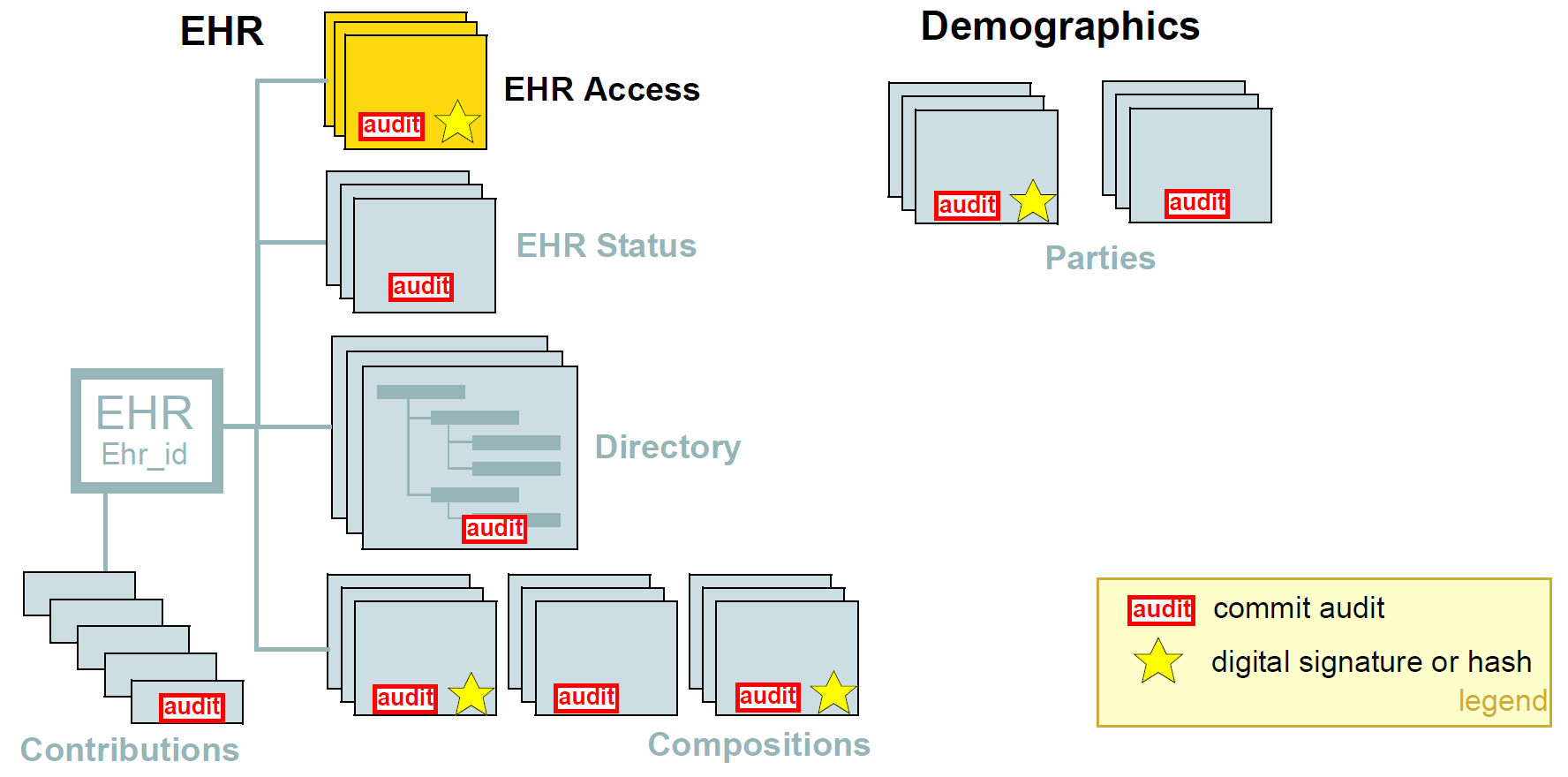
The figure below illustrates the main security measures directly specified by the openEHR architecture. These include EHR/demographic separation and an EHR-wide access control object. At the level of versioned objects, commit audits (mandatory), digital signatures and hashes are available. The following subsections describe these features in more detail.
7.3.2. Security Policy
In and of itself, the openEHR EHR imposes only a minimal security policy profile which could be regarded as necessary, but generally not sufficient for a deployed system (i.e. other aspects would still need to be implemented in layers whose semantics are not defined in openEHR). The following policy principles are embodied in openEHR.
7.3.2.1. General
| Indelibility |
health record information cannot be deleted; logical deletion is achieved by marking the data in such a way as to make it appear deleted (implemented in version control). |
| Audit trailing |
All changes made to the EHR including content objects as well as the EHR status and access control objects are audit-trailed with user identity, time-stamp, reason, optionally digital signature and relevant version information; one exception is where the modifier is the patient, in which case, a symbolic identifier can be used (known as |
| Anonymity |
the content of the health record is separate from identifying demographic information. This can be configured such that theft of the EHR provides no direct clue to the identity of the owning patient (indirect clues are of course harder to control). Stealing an identified EHR involves theft of data from two servers, or even theft of two physical computers, depending on deployment configuration. |
7.3.2.2. Access Control
| Access list |
the overriding principle of access control must be "relevance" both in terms of user identity (who is delivering care to the patient) and time (during the current episode of care, and for some reasonable, limited time afterward). An access control list can be defined for the EHR, indicating both identified individuals and categories, the latter of which might be role types, or particular staff groups. |
| Access control of access settings |
a gate-keeper controls access to the EHR access control settings. The gate-keeper is established at the time of EHR creation as being one of the identities known in the EHR, usually the patient for mentally competent adults, otherwise a parent, legal guardian, advocate or other responsible person. The gate-keeper determines who can make changes to the access control list. All changes to the list are audit-trailed as for normal data (achieved due to normal versioning). |
| Privacy |
patients can mark Compositions in the EHR as having one of a number of levels of privacy. The definition of the privacy levels is not hard-wired in the openEHR models but rather is defined by standards or agreements within jurisdictions of use. |
| Usability |
The general mentality of access control setting is one of "sensible defaults" that work for most of the information in the EHR, most of the time. The defaults for the EHR can be set by the patient, defining access control behaviour for the majority of access decisions. Exceptions to the default policy are then added. This approach minimises the need to think about the security of every item in the EHR individually. |
Other security policy principles that should be implemented in even a minimal EHR deployment but are not directly specified by openEHR include the following.
| Access logging |
read accesses by application users to EHR data should be logged in the EHR system. Currently openEHR does not specify models of such logs, but might do so in the future. Studies have shown that making users aware of the fact of access logging is an effective deterrent to inappropriate access (especially where other controls are not implemented). There are some proponents of the argument that even read-access logs should be made part of the content of the EHR proper; currently openEHR does not support this approach. |
| Record demerging |
when data for a patient is found to be in another patient’s EHR, the access logs for that EHR should be used to determine who has accessed that data, primarily to determine if subsequent clinical thinking (e.g. diagnoses, medication decisions) have been made based on wrong information. |
| Record merging |
when more than one EHR is discovered for the same patient, and have to be merged into a single record, the access control lists have to be re-evaluated and merged by the patient and potentially relevant carers. |
| Time-limitation of access |
mechanisms should be implemented that limit the time during which given health professionals can see the patient record. Usually, the outer limits are defined by the interval of the episode of care in an institution plus some further time to cover follow-up or outpatient care. Episode start and end are recorded in openEHR as instances of the |
| Non-repudiation |
if digital signing of changes to the record is made mandatory, non-repudiation of content can be supported by an openEHR system. The digital signing of communications (EHR Extracts) is also supported in openEHR; coupled with logging of communication of Extracts, this can be used to guarantee non-repudiation of information passed between systems (cf. information passed between back-end and front-end applications of the same system). |
| Certification |
a mechanism should be provided to allow a level of trust to be formally associated with user signing keys. |
A key feature of the policy is that it must scale to distributed environments in which health record information is maintained at multiple provider sites visited by the patient.
As Anderson points out in the BMA study, policy elements are also needed for guarding against users gaining access to massive numbers of EHRs, and inferencing attacks. Currently these are outside the scope of openEHR, and realistically, of most EHR implementations of any kind today.
The following sections describe how openEHR supports the first list of policy objectives.
7.3.3. Integrity
7.3.3.1. Versioning
The most basic security-related feature of openEHR is its support for data integrity. This is mainly
provided by the versioning model, specified in the change_control package in the Common Information
Model, and in the Extract Information Model. Change-set based versioning of all information
in the EHR and demographic services constitutes a basic integrity measure for information, since no
content is ever physically modified, only new versions are created. All logical changes and deletions
as well as additions are therefore physically implemented as new Versions rather than changes to
existing information items. Clearly the integrity of the information will depend on the quality of the
implementation; however, the simplest possible implementations (1 Version = 1 copy) can provide
very good safety due to being write-once systems.
The use of change-sets, known as Contributions in openEHR, provides a further unit of integrity corresponding to all items modified, created or deleted in a single unit of work by a user.
The openEHR versioning model defines audit records for all changed items, which can be basic audits and/or any number of additional digitally signed attestations (e.g. by senior staff). This means that every write access of any kind to any part of an openEHR record is logged with the user identification, time, reason, and potentially other meta-data.
7.3.3.2. Digital Signature
The possibility exists within an openEHR EHR to digitally sign each Version in a Versioned object (i.e. for each Version of any logical item, such as medications list, encounter note etc.). The signature is created as a private-key encryption (e.g. RSA-1) of a hash (e.g. MD5) of a canonical representation (such as in schema-based XML) of the Version being committed. A likely candidate for defining the signature and digest strings in openEHR is the openPGP message format (IETF RFC4880), due to being an open specification and self-describing. The use of RFC 4880 for the format does not imply the use of the PGP distributed certificate infrastructure, or indeed any certification infrastructure; openEHR is agnostic on this point. If no public key or equivalent infrastructure is available, the encryption step might be omitted, resulting in a digest only of the content. The signature is stored within the Version object, allowing it to be conveniently carried within EHR Extracts. The process is shown in the following figure.
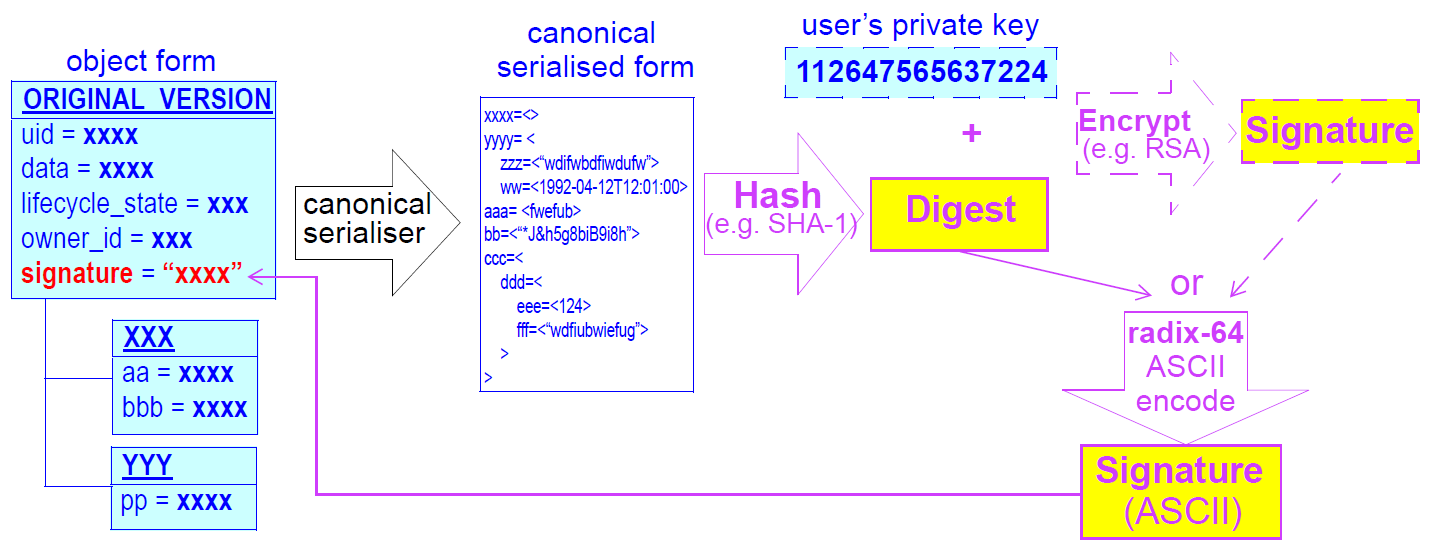
The signing of data in a versioning system acts as an integrity check (the digest performs this function), an authentication measure (the signature performs this function), and also a non-repudiation measure. To guard against hacking of the versioned persistence layer itself, signatures can be forwarded to a trusted notarisation service. A fully secure system based on digital signing also requires certified public keys, which may or may not be available in any given environment.
One of the benefits of digitally signing relatively small pieces of the EHR (single Versions) rather than the whole EHR or large sections of it is that the integrity of items is more immune to localised repository corruptions.
7.3.4. Anonymity
As described above in section 6.1, one of the features of the openEHR EHR is a separation of EHR
(clinical and administrative) information and demographic information. This mainly relates to references
to the patient rather than to provider entities, since the latter are usually publicly known. A special
kind of object known as PARTY_SELF in openEHR is used to refer to the subject in the EHR. The
only information contained in a PARTY_SELF instance is an optional external reference. The
openEHR EHR can be configured to provide 3 levels of separation by controlling whether and where
this external identifier is actually set in PARTY_SELF instances, as follows:
-
Nowhere in the EHR (i.e. every
PARTY_SELFinstance is a blank placeholder). This is the most secure approach, and means that the link between the EHR and the patient has to be done outside the EHR, by associatingEHR.ehr_idand the subject identifier. This approach is more likely for more open environments. -
Once only in the
EHR_STATUSobject (subject attribute), and nowhere else. This is also relatively secure, if the EHR Status object is protected in some way. -
In every instance of
PARTY_SELF; this solution is reasonable in a secure environment, and convenient for copying parts of the record around locally.
This simple mechanism provides a basic protection against certain kinds of information theft or hacking if used properly. In the most secure situation, a hacker has to steal not just EHR data but also separate demographic records and an identity cross reference database, both of which can be located on different machines (making burglary harder). The identity cross-reference database would be easy to encrypt or protect by other security mechanisms.
7.4. Access Control
7.4.1. Overview
Access control is completely specified in an openEHR EHR in the EHR_ACCESS object for the EHR.
This object acts as a gateway for all information access, and any access decision must be made based
on the policies and rules it contains.
One of the problems with defining the semantics of the EHR Access object is that there is currently no published formal, proven model of access control for shared health information. Relevant standards include ISO 13606-4, ISO PMAC (Privilege Management and Access Control) standard. Undoubtedly experimental and even some limited production health information security implementations exist. In reality however, no large-scale shared EHR deployments exist, and so security solutions to date are still developmental.
The openEHR architecture is therefore designed to accommodate alternative models of access control,
each defined by a subtype of the class ACCESS_CONTROL_SETTING (Security IM). This
approach means that a simplistic access control model can be defined and implemented initially, with
more sophisticated models being used later. The "scheme" in use at any given time is always indicated
in the EHR Access object.
8. Versioning
8.1. Overview
Version control is an integral part of the openEHR architecture. An openEHR repository for EHR or
demographic information is managed as a change-controlled collection of "version containers" (modelled
by the VERSIONED_OBJECT<T> class in the common.change_control package), each containing
the versions of a top-level content structure (such as a Composition or Party) as it changes
over time. A version-controlled top-level content structure is visualised below.
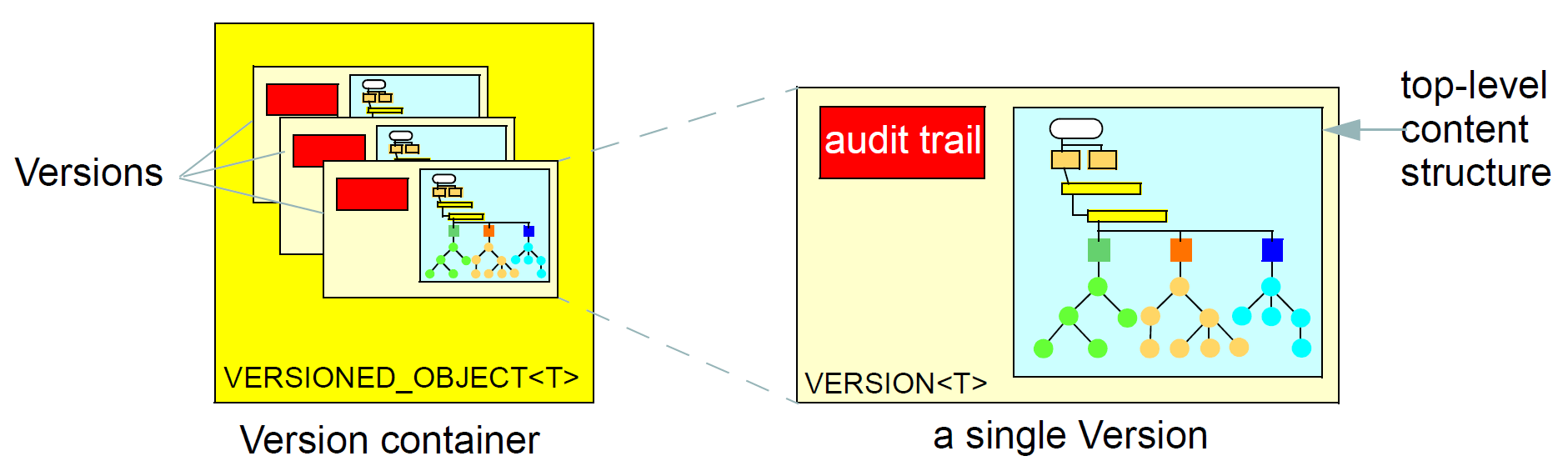
Versioning of single top-level structures is a necessary, but not sufficient requirement for a repository that must provide coherence, traceability, indelibility, rollback, and support for forensic examination of past states of the data. Features supporting "change control" are also required. Under a disciplined change control scheme, changes are not made arbitrarily to single top-level structures, but to the repository itself. Changes take the form of change-sets, called "Contributions", that consist of new or changed versions of the controlled items in the repository. The key feature of a change-set is that it acts like a transaction, and takes the repository from one consistent state to another, whereas arbitrary combinations of changes to single controlled items could easily be inconsistent, and even dangerously wrong where clinical data are concerned.
These concepts are well-known in configuration management (CM), and are used as the basis for most software and other change management systems, including numerous free and commercial products available today. They are a central design feature of openEHR architecture. The following sections provide more details.
8.2. The Configuration Management Paradigm
The configuration management (CM) paradigm is well-known in software engineering, and has its own standard IEEE 828. CM is about managed control of changes to a repository of items (formally called "configuration items" or CIs), and is relevant to any logical repository of distinct information items which changes in time. In health information systems, at least two types of information require such management: electronic health records, and demographic information. In most analyses in the past, the need for change management has been expressed in terms of specific requirements for audit trailing of changes, availability of previous states of the repository and so on. In openEHR, the aim is to provide a formal, general-purpose model for change control, and show how it applies to health information.
8.2.1. Organisation of the Repository
The general organisation of a repository of complex information items such as a software repository, or the EHR consists of the following:
-
a number of distinct information items, or configuration items, each of which is uniquely identified, and may have any amount of internal complexity;
-
optionally, a directory system of some kind, in which the configurations items are organised;
-
other environmental information which may be relevant to correctly interpreting the primary versioned items, e.g. versions of tools used to create them.
In a software or document repository, the CIs are files arranged in the directories of the file system; in an EHR based on openEHR, they are Compositions, the optional Folder structure, Parties in the demographic service and so on. Contributions are made to the repository by users. This general abstraction is visualised as follows.
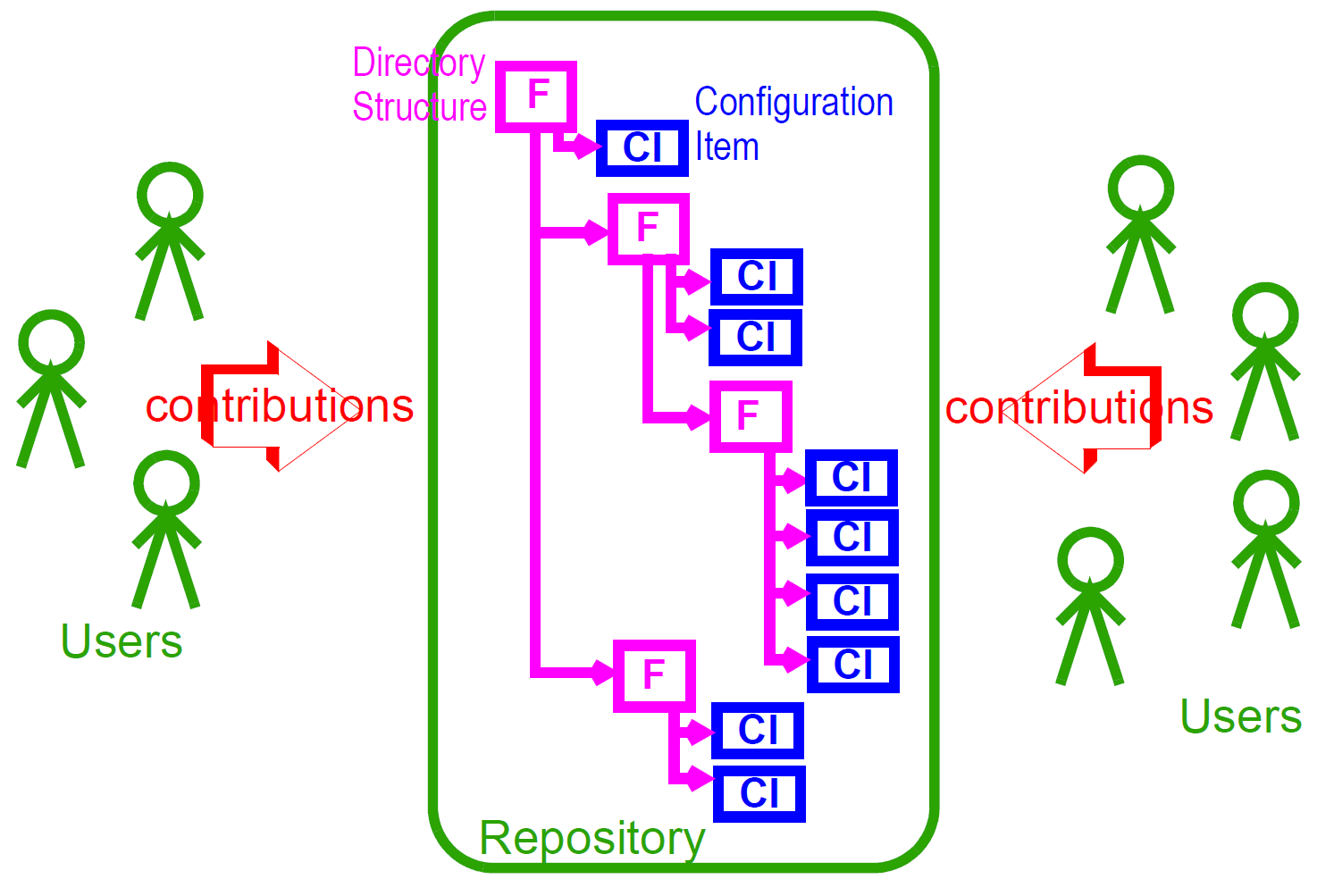
8.2.2. Change Management
Change doesn’t occur to Configuration Items in isolation, but to the repository as a whole. Possible types of change include:
-
creation of a new CI;
-
removal of a CI;
-
modification of a CI;
-
creation of, change to or deletion of part of the directory structure;
-
moving of a CI to another location in the directory structure;
-
attestation of an existing CI.
The goal of configuration management is to ensure the following:
-
the repository is always in a valid state;
-
any previous state of the repository can be reconstructed;
-
all changes are audit-trailed.
8.3. Managing Changes in Time
Properly managing changes to the repository requires two mechanisms. The first, version control, is used to manage versions of each CI, and of the directory structure if there is one. The second is the concept of the "change-set", known as a contribution in openEHR. This is the set of changes to individual CIs (and other top-level structures in the EHR) made by a user as part of some logical change. For example, in a document repository, the logical change might be an update to a document that consists of multiple files (CIs). There is one Contribution, consisting of changes to the document file CIs, to the repository. In the EHR, a Contribution might consist of changes to more than one Composition, and possibly to the organising Folder structure. Any change to the EHR requires a Contribution. The kinds of changes that can occur to items affected in a Contribution are:
-
addition of new item: a new Version container is created and a first Version added to it;
-
deletion of item: a new Version whose data attribute is set to Void is added to an existing Version container;
-
modification of item: a new Version whose data contains the updated form of the item content is added to an existing Version container (this may be done for a logical update or correction);
-
import of item: a new ‘import’ Version is created, incorporating the received Version;
-
attestation of item: a new Attestation is added to the attestations list of an existing Version.
A typical sequence of changes to a repository is illustrated in the following figure.
This shows the effect of four Contributions (left hand side) to a repository containing a number of CIs. As each Contribution is made, the repository is changed in some way. The first brings into existing a new CI, and modifies two others (changes indicated by the 'update' triangles). The second Contribution causes the creation of a new CI due to importing from a lab data feeder system. The third causes a creation as well as three changes, while the fourth causes an amendment to an existing CI.
One nuance which should be pointed out is that in the figure above Contributions are shown as if they are literally a set of deltas, i.e. exactly the changes which occur to the record. Thus, the first Contribution is the set {CIw, Ca1, Cc1, Cd1} and so on. Whether this is literally true depends on the construction of the persistence solution. In some situations, some CIs may be updated by the user viewing the current list and entering just the changes - the situation shown above; in others, the system may provide the current state of these CIs for editing by the user, and submit the updated versions, as shown in the next figure. Some applications may do both, depending on which CI is being updated. The internal versioning implementation may or may not generate deltas as a way of efficient storage.
For the purposes of openEHR, a Contribution is considered as being the set of Versions created or attested at one time, as implied by the figure above.
8.3.1. General Model of a Change-controlled Repository
The following figure shows an abstract model of a change-controlled repository.
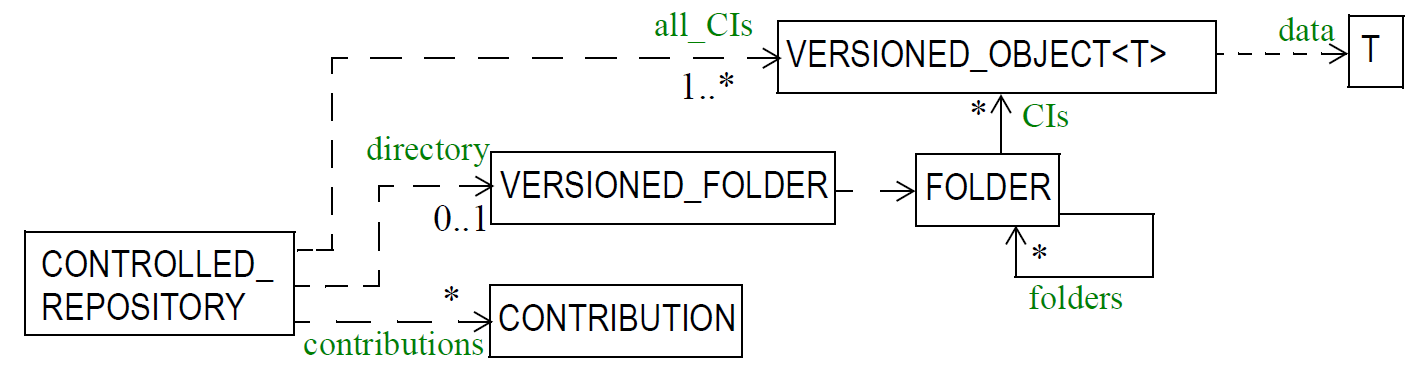
This consists of:
-
version-controlled configuration items - instances of
VERSIONED_OBJECT<T>; -
CONTRIBUTIONs; -
an optional directory system of folders. If folders are used, the folder structure must also be versioned as a unit.
The actual type of links between the controlled repository and the other entities might vary - in some cases it might be association, in others aggregation; cardinalities might also vary. The figure above therefore provides a guide to the definition of actual controlled repositories, such as an EHR, rather than a formal specification for them.
8.4. The Virtual Version Tree
An underlying design concept of the versioning model defined in openEHR is known as a "virtual version tree". The idea is simple in the abstract. Information is committed to a repository (such as an EHR) in lumps, each lump being the "data" of one Version. Each Version has its place within a version tree, which in turn is maintained inside a Versioned object (or "version container"). The virtual version tree concept means that any given Versioned object may have numerous copies in various systems, and that the creation of versions in each is done in such a way that all versions so created are in fact compatible with the "virtual" version tree resulting from the superimposition of the version trees of all copies. This is achieved using simple rules for version identification and is done to facilitate data sharing. Two very common scenarios are served by the virtual version tree concept:
-
longitudinal data that stands as a proxy for the state or situation of the patient such as "Medications" or "Problem list" (persistent Compositions in openEHR) is created and maintained in one or more care delivery organisations, and shared across a larger number of organisations;
-
some EHRs in an EHR server in one location are mirrored into one or more other EHR servers (e.g. at care providers where the relevant patients are also treated); the mirroring process requires asynchronous synchronisation between servers to work seamlessly, regardless of the location, time, or author of any data created.
The versioning scheme used in openEHR guarantees that no matter where data are created or copied, there are no inconsistencies due to sharing, and that logical copies are explicitly represented. It therefore provides direct support for shared data in a shared care context.
9. Identification
9.1. Identification of the EHR
In an openEHR system, each EHR has a unique identifier, known as the EHR id, found in the root EHR object of each EHR. EHR ids should be "strong" globally unique identifiers such as reliably created Oids or Guids. No single system should contain two EHRs for the same subject. If this is not the case, it means that the EHR system has failed to detect the existence of an EHR for a subject, or failed to match provided demographic attributes to the subject.
In a distributed environment, the correspondence of EHR ids to subjects (i.e. patients) is variable, and depends on the level of integration of the environment. In non-integrated or sporadically connected environments, the same patient is likely to have a separate EHR at each institution, each with its own unique EHR id, but the same subject id. If copies of parts of the patient EHR at one location is requested by another provider, the received items will be merged into the local EHR for that patient. Merges of persistent Compositions in such circumstances are likely to require human intervention. Multiple EHR ids per patient in a distributed context are evidence of a lack of systematic connectivity or identification service.
In a fully integrated distributed environment, the typical patient will still have local EHRs in multiple locations, but each carrying the same EHR id. When a patient presents in a new location, a request to the environment’s identification service could be made to determine if there is already an EHR for this patient. If there is, a clone of all or part of the existing EHR could be made, or a new empty EHR could be created, but in all cases, the EHR id would be the same as that used in other locations for the same patient.
Note that the above logic only holds where the EHR in each location is an openEHR EHR.
9.2. Identification of Items within the EHR
9.2.1. General Scheme
While identification of EHRs is not completely definable by openEHR, the identification of items within an EHR is fully defined. The scheme described here requires two kinds of "identifier": identifiers proper and references, or locators. An identifier is a unique (within some context) symbol or number given to an object, and usually written into the object, whereas a reference is the use of an identifier by an exterior object, to refer to the object containing the identifier in question. This distinction is the same as that between primary and foreign keys in a relational database system.
In the openEHR RM, identifiers and references are implemented with two groups of classes defined
in the support.identification package. Identifiers of various kinds are defined by descendant
classes of OBJECT_ID, while references are defined by the classes inheriting from OBJECT_REF. The
distinction is illustrated in the following figure. Here we see two container objects with OBJECT_IDs (since
OBJECT_ID is an abstract type, the actual type will be another XXX_ID class), and various
OBJECT_REFs as references.
9.2.2. Levels of Identification
In order to make data items locatable from the outside, identification is supported at 3 levels in openEHR, as follows:
-
version containers:
VERSIONED_OBJECTs(Common IM) are identified uniquely; -
top-level content structures: content structures such as
COMPOSITION,EHR_STATUS,EHR_ACCESS,PARTYetc. are uniquely identified by the association of the identifier of their containingVERSIONED_OBJECTand the identifier of their containingVERSIONwithin the container; -
internal nodes: nodes within top-level structures are identified using paths.
Three kinds of identification are used respectively. For version containers, meaningless unique identifiers (UIDs) are used. In most cases, the type HIER_OBJECT_ID will be used, which contains an instance of a subtype of the UID class, i.e. either an ISO OID or a UUID (IETF RFC4122; also known as a GUID). UUIDs are strongly preferred in openEHR since they require no central assignment and can be generated on the spot. A versioned container can be then referenced with an OBJECT_REF containing its identifier.
Versions of top-level structures (i.e. objects of type VERSION<X>, such as VERSION<COMPOSITION>) are identified in a way that is guaranteed to work even in distributed environments where copying, merging and subsequent modification occur. The full identification of a Version is the globally unique tuple consisting of the uid of the owning VERSIONED_OBJECT, and the two VERSION attributes creating_system_id and version_tree_id. The
creating_system_id attribute carries a unique identifier for the system where the content was first created; this may be a GUID, Oid or reverse internet identifier. The version_tree_id is a 1 or 3-part number string, such as "1" or for a branch, "1.2.1". A typical version identification tuple is as follows:
F7C5C7B7-75DB-4b39-9A1E-C0BA9BFDBDEC -- id of VERSIONED_COMPOSITION
au.gov.health.rdh.ehr1 -- id of creating system
2 -- current versionThis 3-part tuple is known as a version locator and is defined by the class OBJECT_VERSION_ID in the identification package section of the Base Types specification. The openEHR version identification scheme is described in detail in the change_control package section of the Common IM.
The contained top-level content item (i.e. a COMPOSITION etc) also has a uid attribute (inherited from LOCATABLE), and it is strongly recommended that this be populated with a copy of the OBJECT_VERSION_ID from the containing VERSION<X> object. This facilitates identifying versions from a naked content object e.g. returned in a query.
A VERSION can be referred to using a normal OBJECT_REF that contains a copy of the version’s OBJECT_VERSION_ID.
The last component of identification is the path, used to refer to an interior node of a top-level structure of some type X (e.g. COMPOSITION, PARTY, etc), the latter identified by its Version locator. Paths in openEHR follow an Xpath style syntax, with slight abbreviations to shorten paths in the most common cases. Paths are described in detail below.
To refer to an interior data node from outside a top-level structure, a combination of a version locator
and a path is required. This is formalised by the LOCATABLE_REF class, also in the identification
package section of the Base Types specification. A Universal Resource Identifier (URI) form can also be used,
defined by the data type DV_EHR_URI. This type provides a single string expression
in the scheme-space ehr: which can be used to refer to an interior data node from anywhere. (It can
also be used to represent queries; see below). Any LOCATABLE_REF can be converted to a
DV_EHR_URI, although not all DV_EHR_URIs are LOCATABLE_REFs.
The figure below summarises how various types of OBJECT_ID and OBJECT_REF are used to identify
objects, and to reference them from the outside, respectively.
10. Archetypes and Templates
10.1. Overview
Under the multi-level modelling approach, the formal definition of information structuring occurs at two levels. The lower level is that of the reference model, a stable object model from which software and data can be built. Concepts in the openEHR reference model are invariant, and include things like Composition, Section, Observation, and various data types such as Quantity and Coded text. The upper level consists of domain-level definitions in the form of archetypes and templates. Concepts defined at this level include things such as 'blood pressure measurement', 'SOAP headings', and 'HbA1c Result'.
All information conforming to the openEHR Reference Model (RM) - i.e. the collection of Information Models (IMs) - is 'archetypable', meaning that the creation and modification of the content, and subsequent querying of data is controllable by archetypes. Archetypes are themselves separate from the data, and are stored in their own repository. The archetype repository at any particular location will usually include archetypes from well-known online archetype libraries. Archetypes are deployed at runtime via templates that specify particular groups of archetypes to use for a particular purpose, often corresponding to a screen form.
Archetypes are themselves instances of an archetype model, which defines a language in which to write archetypes; the syntax equivalent of the model is the Archetype Definition Language, ADL. These formalisms are specified in the openEHR Archetype Object Model (AOM) and ADL documents respectively. Each archetype is a set of constraints on the reference model, defining a subset of instances that are considered to conform to the subject of the archetype, e.g. 'laboratory result'. An archetype can thus be thought of as being similar to a LEGO® instruction sheet (e.g. for a tractor) that defines the configuration of LEGO® bricks making up a tractor. Archetypes are flexible; one archetype includes many variations, in the same way that a LEGO® instruction might include a number of options for the same basic object. Mathematically, an archetype is equivalent to a query in F-logic Kifer Lausen & Wu (2000).
In terms of scope, archetypes are general-purpose, re-usable, and composable. For data capture and validation purposes, they are usually used at runtime by templates. An openEHR Template is a specification that defines a tree of one or more archetypes, each constraining instances of various reference model types, such as Composition, Section, Entry subtypes and so on. Thus, while there are likely to be archetypes for such things as 'biochemistry results' (an Observation archetype) and 'SOAP headings' (a Section archetype), templates are used to put archetypes together to form whole Compositions in the EHR, e.g. for 'discharge summary', 'antenatal exam' and so on. Templates usually correspond closely to screen forms, printed reports, and in general, complete application-level lumps of information to be captured or sent; they may therefore be used to define message content. They are generally developed and used locally, while archetypes are usually widely used.
A template is used at runtime to create default data structures and to validate data input, ensuring that all data in the EHR conform to the constraints defined in the archetypes referenced by the template. In particular, it conforms to the path structure of the archetypes, as well as their terminological constraints. Which archetypes were used at data creation time is written into the data, in the form of both archetype identifiers at the relevant root nodes, and archetype node identifiers (the [atnnnn] codes), which act as normative node names, and which are in turn the basis for paths. When it comes time to modify the same data, these archetype node identifiers enable applications to retrieve and use the original archetypes, ensuring modifications respect the original constraints.
Archetypes also form the basis of semantic querying. Queries are expressed in a language which is a synthesis of SQL (SELECT/FROM/WHERE) and W3C XPaths, extracted from the archetypes.
10.2. Archetype Formalisms and Models
10.2.1. Overview
In openEHR, archetypes are formalised by the Archetype Object Model (AOM). This is an object model of the semantics of archetypes. When an archetype is represented in memory (for example in an archetype-enabled EHR 'kernel'), the archetype will exist as instances of the classes of this model. The AOM is thus the definitive statement of the semantics of archetypes.
In serialised form, archetypes can be represented in various ways. The normative, abstract serialisation in openEHR is Archetype Definition Language (ADL). This is an abstract language based on Frame Logic queries with the addition of terminology. An ADL archetype is a guaranteed 100% lossless rendering of the semantics of any archetype, and is designed to be a syntactic analogue of the AOM. Nevertheless, other lossless and lossy serialisations are possible and some already exist. For practical purposes, XML-based serialisations are used in some situations. A serialisation purely expressed in ODIN, the ADL object serialisation syntax is available for ADL2 archetypes. Various HTML, RTF and other formats are used for screen rendering and human review.
openEHR ADL2 Templates are represented as ODIN documents whose object model conforms to the AOM.
10.2.2. Design-time Relationships between Archetypes
Archetypes are extensible formal constraint definitions of object structures. In common with object
model classes, they can be specialised, as well as composed (i.e. aggregated). Specialised archetypes
are created when an archetype is already available for the content that needs to be modelled, but it
lacks detail or is too general. For example, the archetype openEHR-EHR-OBSERVATION.laboratory.v1
contains generic concepts of 'specimen', 'diagnostic service', a single result of any type,
and a two-level result battery for grouped results. This archetype could be (and has been) used to represent
nearly any kind of laboratory result data. However, specialisations such as openEHR-EHROBSERVATION.laboratory-glucose.v1
are extremely useful, and can be easily defined based
on the predecessor; in this case, the single result node is redefined to be 'blood glucose'. The formal
rule for specialisation is that:
-
a specialised archetype can only further narrow existing constraints in the parent (but it may add its own).
This has the effect that the data created with any specialised archetype will always be matched by queries based on the parent archetype - in other words, a query for 'laboratory' Observations will correctly retrieve 'glucose' Observations as well. This accords with the basic ontological principle of subsumption, which says that instances of a type B are also instances of type A, where type B is related to type A by the semantic relationship 'IS-A'. Specialised archetypes are indicated by the use of an identifier derived from the parent archetype, with a new sub-element of the semantic part of the identifier, separated by a '-' character.
The second relationship possible between archetypes is composition, allowing large data structures to
be flexibly constrained via the hierarchical re-use of smaller archetypes. Composition is defined in
terms of 'slots' within an archetype. A slot is a point in an archetype structure where, instead of specifying
an object type inline, a special allow_archetype constraint is used to specify other archetypes
constraining that same type, that may be used at that point. For example, the archetype openEHR-EHR-SECTION.vital_signs.v1
defines a heading structure for headings to do with vital signs. It
also defines as its items attribute value (i.e. what comes under the heading) a number of possible
Observations; however, rather than defining these inline, it specifies an archetype slot in the form of
constraints on Observation archetypes that are allowed at that point. The simplest kind of constraint is
in terms of regular expressions on archetype identifiers. More complex constraints can be stated in
terms of paths in other archetypes (for example exists (/some/path[at0005])). A slot thus
defines a 'chaining point' in terms of possible archetypes allowed or excluded at that point; limiting
this to a single archetype is of course possible. Templates are used to choose which particular archetypes
allowed at a slot will actually be used in a given circumstance.
10.3. Relationship of Archetypes and Templates to Data
All nodes within the top-level information structures in the openEHR RM are 'archetypable', with
certain nodes within those structures being archetype 'root points'. Each top-level type is always
guaranteed to be an archetype root point. Although it is theoretically possible to use a single archetype
for an entire top-level structure, in most cases, particularly for COMPOSITION and PARTY, a hierarchical
structure of multiple archetypes will be used, via the slot mechanism described above. This
allows for componentisation and reusability of archetypes. When hierarchies of archetypes are used
for a top-level structure, there will also be archetype root points in the interior of the structure. For
example, within a COMPOSITION, ENTRY instances (i.e. OBSERVATIONs, EVALUATIONs etc.) are
almost always root points. SECTION instances are root points if they are the top instance in a Section
structure; similarly for FOLDER instances within a directory structure. Other nodes (e.g. interior SECTIONs,
ITEM_STRUCTURE instances) might also be archetype root points, depending on how archetypes
are applied at runtime to data. The following figure illustrates the application of archetypes and
templates to data.

10.4. Archetype-enabling of Reference Model Data
Archetype-enabling of Reference Model classes is achieved via inheritance of the class LOCATABLE
from the package common.archetyped (see Common IM). The LOCATABLE class includes the
attributes archetype_node_id and archetype_details. In the data, the former carries an identifier from
the archetype. If the node in the data is a root point, it carries the multipart identifier of the generating
archetype, and archetype_details carries an ARCHETYPED object, containing information pertinent to
archetype root points. If it is a non-root node, the archetype_node_id attribute carries the identifier
(known as an 'at', or 'archetype term' code) of the archetype interior node that generated the data
node, and the archetype_details attribute is void.
Sibling nodes in data can carry the same archetype_node_id in some cases, since archetypes provide a
pattern for data, rather than an exact template. In other words, depending on the archetype design, a
single archetype node may be replicated in the data.
In this way, each archetyped data composition in openEHR data has a generating archetype which
defines the particular configuration of instances to create the desired composition. An archetype for
'biochemistry results' is an OBSERVATION archetype, and constrains the particular arrangement of
instances beneath an OBSERVATION object; a 'problem/SOAP headings' archetype constrains SECTION
objects forming a SOAP headings structure. In general, an archetyped data composition is any
composition of data starting at a root node and continuing to its leaf nodes, at which point lower-level
compositions, if they exist, begin. Each of the archetyped areas and its subordinate archetyped areas in Figure 36 is an archetyped data composition.
|
Note
|
care must be taken not to confuse the general term 'composition' with the specific use of this word in
openEHR and ISO 13606, defined by the COMPOSITION class; the specific use is always indicated by
using the term 'Composition'.
|
The result of the use of archetypes to create data in the EHR (and other systems) is that the structure of data in any top-level object conforms to the constraints defined in a composition of archetypes chosen by a template, including all optionality, value, and terminology constraints.
10.5. Archetypes, Templates and Paths
The use of openEHR archetypes and templates enables paths to be used ubiquitously in the openEHR
architecture. Paths are extracted from Archetypes and templates, and are constructed from attribute
names and archetype node identifiers, in an Xpath-compatible syntax, as shown in the following figure.
These paths serve to identify any node in a template or archetype, such as the 'diastolic blood pressure'
ELEMENT node, deep within a 'blood pressure measurement' archetype. Since archetype node
identifiers are embedded into data at runtime, archetype paths can be used to extract data nodes conforming
to particular parts of archetypes, providing a very powerful basis for querying. Paths can also
be constructed into data, using more complex predicates (still in the Xpath style). Paths in openEHR
are explained in detail under Section 11.
10.6. Archetypes and Templates at Runtime
10.6.1. Overview
openEHR archetypes and templates were designed as formal artefacts, so as to be computable at runtime. They perform two key functions. The first is to facilitate data validation at data capture or import time, i.e. to guarantee that data conform to not just the reference model, but also to the archetypes themselves. Data validation with archetypes is mediated by the use of openEHR Templates. The second function is as a design basis for queries. Since data are captured based on archetypes, all openEHR data are guaranteed to conform to the 'semantic paths' that are created by the composition of archetypes within a template. The paths (such as those shown in figure Figure 37 above) are incorporated within a familiar SQL-style syntax, to form queries that can be evaluated to retrieve items on a semantic basis.
10.6.2. Deploying Archetypes and Templates
Archetypes are mostly designed by clinical or other domain experts, and often require significant study of a subject area, for example, obstetrics. The development process may occur at a national or international level, and requires peer review and testing in real systems. This accords with the semantic value of archetypes, namely as reusable models of content. Consequently, from the point of view of any given site of deployment, archetypes are most likely to have been developed elsewhere, and to reside in a recognised, quality assured repository.
Such a repository may contain hundreds or even thousands of archetypes. However, most EHR sites will only require a relatively small number. Clinical experts estimate that 100 archetypes would take care of 80% of routine general practice and acute care, including laboratory, with many of these being specialisations of a much smaller number of key archetypes. However, which 100 archetypes are useful for a given site may well vary based on the kind of health care provided, e.g. diabetic clinic, cancer, orthopedic hospital ward, aged care home. In general, it can be expected that nearly all archetype deployment sites will use only a small percentage of published archetypes. Some sites may also develop a small number of their own archetypes; invariably these will be specialisations of existing archetypes.
While archetypes constitute the main shared and carefully quality-assured design activity in the second layer of openEHR’s two-level structure, templates are a more local affair, and are likely to be the point of contact of many system designers with archetypes. A template will typically be designed based on three things:
-
what is desired to be in a screen form or report;
-
what archetypes are already available;
-
local usage of terminology.
Templates will generally be created locally by tools conforming to the openEHR Template Object Model.
In the case of GUI applications, the final step in the chain is GUI screen forms. These are created in a multitude of ways and technologies. In some cases, they will be partially or completely generated from templates. Regardless of the details, the connection between a screen form and a template will be established in the tooling environment, so that when the form is requested by a user, the relevant template will be activated, in turn activating the relevant archetypes.
A further technical detail may come into play in many deployment situations: since the archetypes and templates required by the environment will be known in advance, they may well be compiled into a near-runtime form from the sharable openEHR form (i.e. ADL, TOM files) in which they are received from a repository or local tool. This form will usually differ from site to site, and both improves performance and ensures that only validated archetypes and templates will actually be accessed by applications. In such systems, runtime form of templates is most likely to incorporate copies of the relevant archetypes.
The deployment of archetypes, templates, and screen forms is shown below.
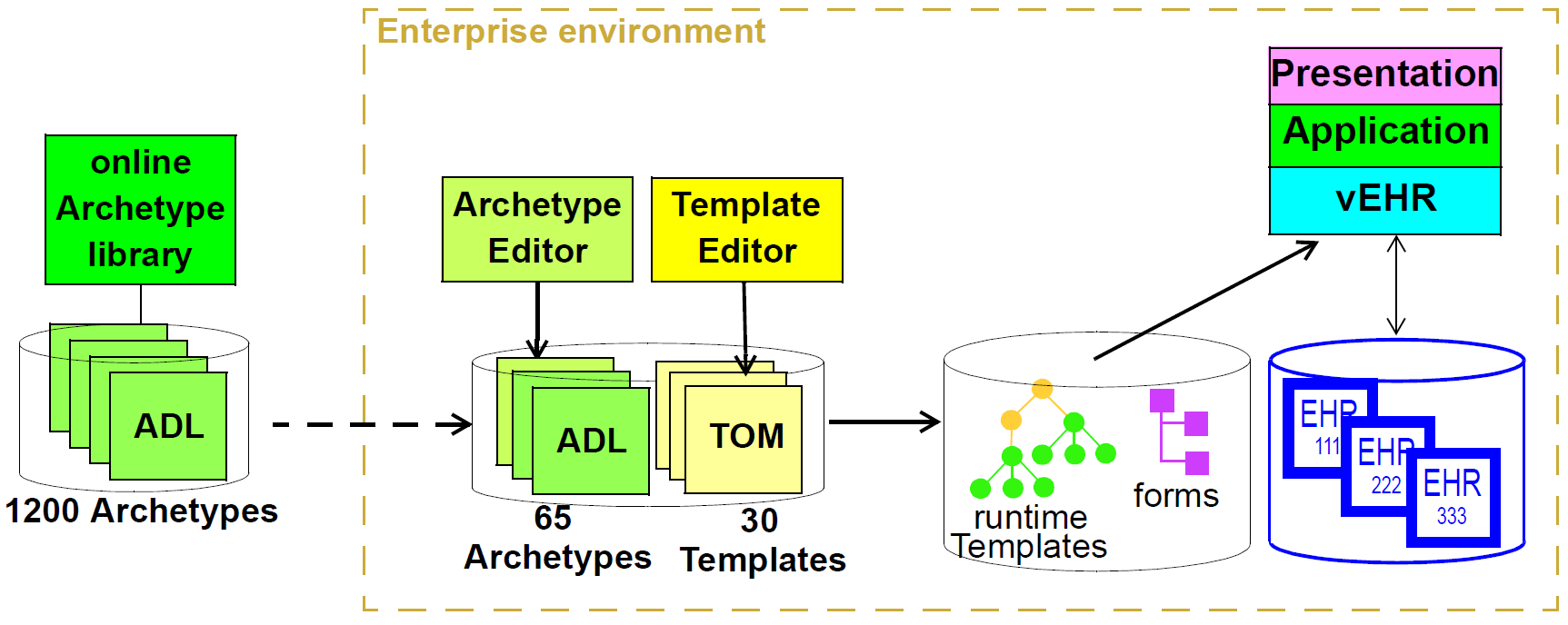
10.6.3. Validation during Data Capture
Validation is the primary runtime function of archetypes - it is how 'archetype-based' data are created in the first place, and modified thereafter. Archetype-based validation can be used in a GUI application or in a data import service. Although the source of the data (keystrokes or received XML or other messages) is different, the logical process is the same: create archetype-based openEHR data according to the input stream.
The process at runtime may vary in some details according to implementations and other aspects of the care setting, but the main thrust will be the same. The archetypes used at a particular site will always be mediated at runtime by openEHR templates developed for that site or system; these will usually be linked to screen forms or other formal artefacts that enable the connection between archetypes and the user or application. It will not be uncommon for a template to be constructed partially at runtime, due to user choices of archetypes being made on the screen, although of course the user will not be directly aware of this. Regardless, by the time data are created and validated against the relevant archetypes, the template that does the job will be completely specified.
The actual process of data creation and committal is illustrated below. The essence of the process is that a 'kernel' component performs the task of data creation and validation by maintaining a 'template space' and a 'data space'. The former contains the template and archetypes retrieved due to a screen form being displayed; the latter contains the data structures (instances of the openEHR reference model) that are constructed due to user activity on the screen. When data are finally committed, they are guaranteed to conform to the template/archetype definitions, due to the checks that are made each time the user tries to change the data structure. The committed data contain a 'semantic imprint' of the generating archetypes, in the form of archetype node identifiers on every node of the data. This simple inclusion in the data model ensures that all archetypes data are queryable by the use of archetype paths. In XML representations, the archetype node ids are represented as XML attributes (i.e. inside the tag), thus enabling XPaths to be conveniently navigated through the data based on these identifiers (more details on this are in the next section).
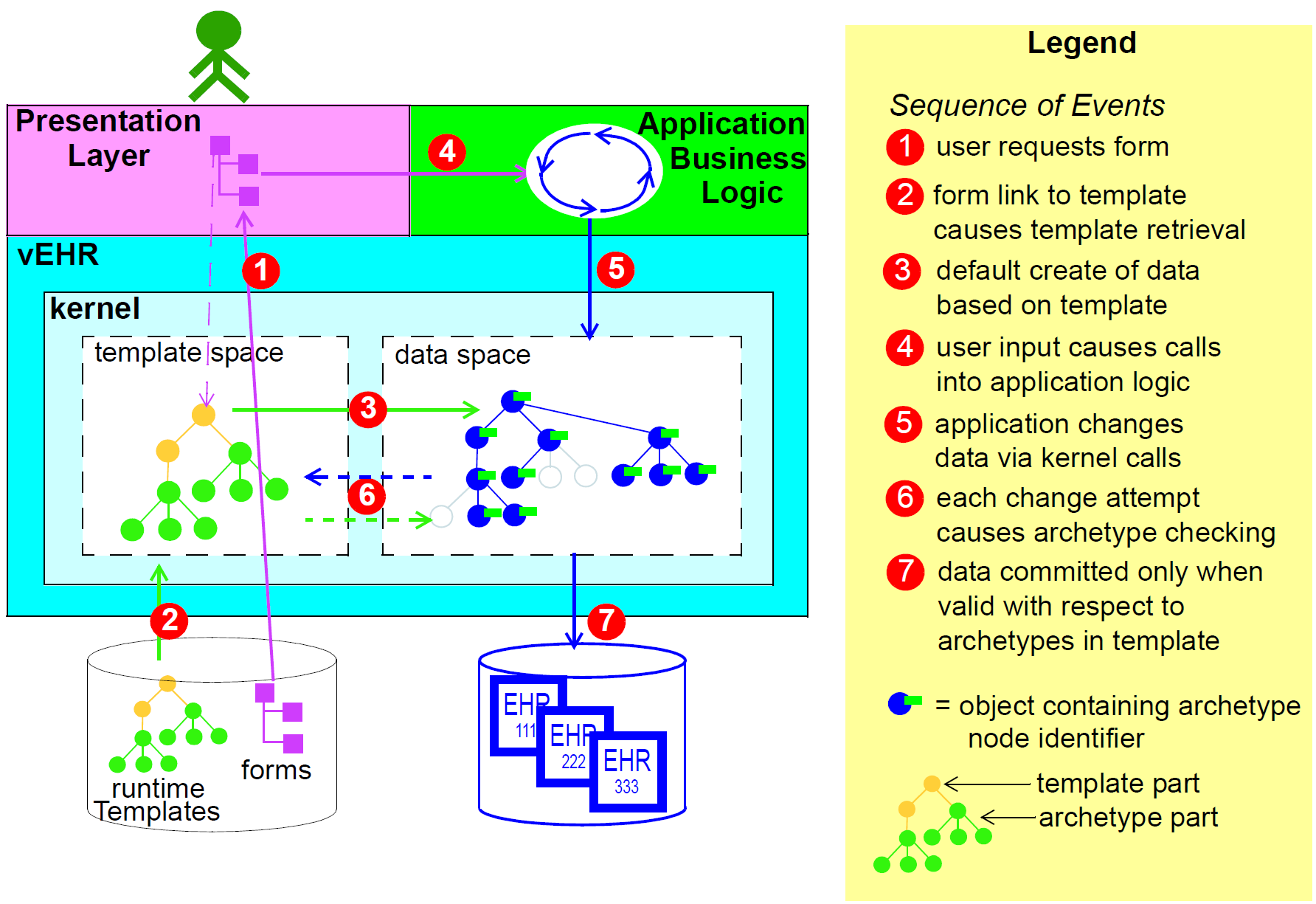
If data are later modified, they are brought into the kernel along with the relevant template and archetypes, and the embedded node identifiers allow the kernel to continue to perform appropriate checking of changes to the data.
10.6.4. Querying
The second major computational function of archetypes is to support querying. As described above, and in the next section, the paths extracted from archetypes are the basis for queries into the data. Queries are defined in AQL (Archetype Query Language), which is essentially a synthesis of SQL and XPath style paths extracted from archetypes. The following is an example AQL query meaning "Get the BMI values which are more than 30 kg/m2 for a specific patient":
SELECT o/[at0000]/data[at0001]/events[at0002]/data[at0003]/item[0004]/value
FROM EHR [uid=@ehrUid]
CONTAINS COMPOSITION c [openEHR-EHR-COMPOSITION.report.v1]
CONTAINS OBSERVATION o[openEHR-EHR-OBSERVATION.body_mass_index.v1]
WHERE o/[at0000]/data[at0001]/events[at0002]/data[at0003]/item[0004]/value > 3010.7. The openEHR Archetypes
A set of heavily reviewed archetypes is available on the openEHR Clinical Knowledge Manager (CKM). This collection is authored by hundreds of clinical professionals, and is constantly growing.
11. Paths and Locators
11.1. Overview
The openEHR architecture includes a path mechanism that enables any node within a top level structure to be specified from the top of the structure using a "semantic" (i.e. archetype-based) X-path compatible path. The availability of such paths radically changes the available querying possibilities with health information, and is one of the major distinguishing features of openEHR.
Technically, the combination of a path and a Version identifier such as OBJECT_VERSION_ID forms
a 'globally qualified node reference' which can be expressed using LOCATABLE_REF. It can also be
expressed in portable URI form as a DV_EHR_URI, known as a 'globally qualified node locator'.
Either representation enables any openEHR data node to be referred to from anywhere. This section
describes the syntax and semantics of paths, and of the URI form of reference. In the following, the
term 'archetype path' means a path extracted from an archetype, while "data path" means one that
identifies an item in data. They are no different formally, and this terminology is only used to indicate
where they are used.
11.2. Paths
11.2.1. Basic Syntax
Paths in openEHR are defined in an Xpath1-compatible syntax which is a superset of the path syntax described in the Archetype Definition Language (ADL). The syntax is designed to be easily mappable to W3C Xpath expressions, for use with openEHR-based XML.
The data path syntax used in locator expressions follows the general pattern of a path consisting of segments each consisting of an attribute name, and separated by the slash ('/') character, i.e.:
attribute_name / attribute_name / ... / attribute_name
|
Note
|
In all openEHR documentation, the term 'attribute' is used in the object-oriented sense of 'property of an object', not in the XML sense of named values appearing within a tag. The syntax described here should not be considered to necessarily have a literal mapping to XML instance, but rather to have a logical mapping to object-oriented data structures. |
Paths select the object which is the value of the final attribute name in the path, when going from some starting point in the tree and following attribute names given in the path. The starting point is indicated by the initial part of the path, and can be specified in two ways:
-
relative path: path starts with an attribute name, and the starting point is the current point in the tree (given by some previous operation or knowledge);
-
absolute path: path starts with a
/; the starting point is the top of the structure.
In addition, the // notation from Xpath can be used to define a path pattern:
-
path pattern: path starts with or contains the symbol '//' and is taken to be a pattern which can match any number of path segments in the data; the pattern is matched if an actual path can be found anywhere in the structure for which part of the path matches the path section before the
//symbol, and a later section matches the section appearing after the//.
11.2.2. Predicate Expressions
11.2.2.1. Overview
Paths specified solely with attribute names are limited in two ways. Firstly, they can only locate objects in structures in which there are no containers such as lists or sets. However, in any realistic data, including most openEHR data, list, set and hash structures are common. Additional syntax is needed to match a particular object from among the siblings referred to by a container attribute. This takes the form of a predicate expression enclosed in brackets ('[]') after the relevant attribute in a segment, i.e.:
attribute_name [predicate expression]
The general form of a path then resembles the following:
attribute_name / attribute_name [predicate expression] / ...
Here, predicate expressions are used optionally on those attributes defined in the reference model to be of a container type (i.e. having a cardinality of > 1). If a predicate expression is not used on a container attribute, the whole container is selected. Note that predicate expressions are often possible even on single-valued attributes, and can be used (e.g. if generic path-processing software can’t tell the difference) but are not required.
The second limitation of basic paths is that they cannot locate objects based on other conditions, such as the object having a child node with a particular value. To address this, predicate expressions can be used to select an object on the basis of other conditions relative to the object, by including boolean expressions including paths, operators, values and parentheses. The syntax of predicate expressions used in openEHR is a subset of the Xpath syntax for predicates with a small number of short-cuts.
11.2.2.2. Archetype path Predicate
The most important predicate uses the archetype_node_id value (inherited from LOCATABLE) to limit the items returned from a container, such as to certain ELEMENTs within a CLUSTER. The shortcut form allows the archetype code to be included on its own as the predicate, e.g. [at0003]. This shortcut corresponds to using an archetype path against the runtime data. A typical archetype-derived path is the following (applied to an Observation instance):
/data/events[at0003]/data/items[at0025]/value/magnitude
This path refers to the magnitude of a 1-minute Apgar total in an Observation containing a full Apgar result structure. In this path, the [atNNNN] predicates are a shortcut for [@archetype_node_id = 'atNNNN'] in standard Xpath.
|
Note
|
while an archetype path is always unique in an archetype, it can correspond to more than one item in runtime data, due to the repeated use of the same archetype node within a container. |
11.2.2.3. Name-based Predicate
If the LOCATABLE.name value in certain data is populated with meaningful values, a useful predicate can be formed using a combination of name.value (which takes the Xpath-like form name/value in a predicate) and the archetype_node_id value. The standard Xpath form of this expression is exemplified by the following:
/data/events[@archetype_node_id = 'at0001' and name/value='standing']
with the openEHR equivalent being:
/data/events[at0001 and name/value='standing']
Since the combination of an archetype node identifier and a name value is very common in archetyped databases, a shortcut is also available for the name/value expression, which is to simply include the value after a comma as follows:
/data/events[at0001, 'standing']
11.2.2.4. Other Predicates
Other predicates can be used, based on the value of other attributes such as ELEMENT.name or
EVENT.time. Combinations of the archetype_node_id and other such values are commonly used in
querying, such as the following path fragment (applied to an OBSERVATION instance):
/data/events[at0007 AND time >= '24-06-2005T09:30:00']
This path would choose Events in OBSERVATION.data whose archetype_node_id meaning is 'summary
event' (at0007 in some archetype) and which occurred at or after the given time. The following
example would choose an Evaluation containing a diagnosis (at0002.1) of 'other bacterial intestinal
infections' (ICD10 code A04):
/data/items[at0002.1
AND value/defining_code/terminology_id/value = 'ICD10AM'
AND value/defining_code/code_string = 'A04']
11.2.3. Paths within Top-level Structures
Paths within top-level structures strictly adhere to attribute and function names in the relevant parts of the reference model. Predicate expressions are needed to distinguish multiple siblings in various points in paths into these structures, but particularly at archetype "chaining" points. A chaining point is where one archetype takes over from another as illustrated in figure Figure 36. Chaining points in Compositions occur between the Composition and a Section structure, potentially between a Section structure and other sub-Section structures (constrained by a different Section archetype), and between either Compositions or Section structures, and Entries. Chaining might also occur inside an Entry, if archetyping is used on lower level structures such as Item_lists etc. Most chaining points correspond to container types such as List<T> etc., e.g. COMPOSITION.content is defined to be a List<CONTENT_ITEM>, meaning that in real data, the content of a Composition could be a List of Section structures. To distinguish between such sibling structures, predicate expressions are used, based on the archetype_id. At the root point of an archetype in data (e.g. top of a Section structure), the archetype_id carries the identifier of the archetype used to create that structure, in the same manner as any interior point in an archetyped structure has an archetype_node_id attribute carrying archetype node_id values. The chaining point between Sections and Entries works in the same manner, and since multiple Entries can occur under a single Section, archetype_id predicates are also used to distinguish them. The same shorthand is used for archetype_id predicate expressions as for archetype_node_ids, i.e. instead of using [@archetype_id = "xxxxx"], [xxxx] can be used instead.
The following paths are examples of referring to items within a Composition:
/content[openEHR-EHR-SECTION.vital_signs.v1 and name/value='Vital signs']/items[openEHR-EHR-OBSERVATION.heart_rate-pulse.v1 and name/value='Pulse']/data/events[at0003 and name/value='Any event']/data/items[at1005] /content[openEHR-EHR-SECTION.vital_signs.v1 and name/value='Vital signs']/items[openEHR-EHR-OBSERVATION.blood_pressure.v1 and name/value='Blood pressure']/data/events[at0006 and name/value='any event']/data/items[at0004] /content[openEHR-EHR-SECTION.vital_signs.v1, 'Vital signs']/items[openEHR-EHR-OBSERVATION.blood_pressure.v1, 'Blood pressure']/data/events[at0006, 'any event']/data/items[at0005]
Paths within the other top level types follow the same general approach, i.e. are created by following the required attributes down the hierarchy.
11.2.4. Data Paths and Uniqueness
Archetype paths are not guaranteed to uniquely identify items in data, due to the fact that one archetype
node may correspond to multiple instances in the data. However it is often useful to be able
to construct a unique path to an item in real data. This can be done by using attributes other than
archetype_node_id in path predicates.
11.2.4.1. Using a Uid-based Predicate
The most reliable way to obtain unique path for run-time nodes in data is is by populating the inherited LOCATABLE.uid field with UUIDs. A predicate can be formed from just the uid value, or the combination of uid value and the archetype_node_id value, which although technically speaking is redundant, is more informative (e.g. it can be displayed with the archetype_node_id meaning visible for the user). This is the preferred method to achieve runtime unique node identification. The standard Xpath form of this expression is exemplified by the following:
/data/events[@uid='25f2f224-64f0-41ec-a5c7-c31c040c77ce'] <!-- assumes 'uid' is an XML attribute in XSD --> /data/events[@archetype_node_id = 'at0001' and @uid='25f2f224-64f0-41ec-a5c7-c31c040c77ce']
with the openEHR equivalent being:
/data/events[uid='25f2f224-64f0-41ec-a5c7-c31c040c77ce'] /data/events[at0001 and uid='25f2f224-64f0-41ec-a5c7-c31c040c77ce']
11.2.4.2. Using a Name-based Predicate
If the LOCATABLE.name value in certain data is known to be reliably populated with unique values across immediate siblings, the name/value term may be used as described above to form a uniquely identifying predicate for a node. Consider as an example the following OBSERVATION archetype (expressed in ODIN syntax):
OBSERVATION[at0000] matches { -- blood pressure measurement
data matches {
HISTORY matches {
events {1..*} matches {
EVENT[at0006] {0..1} matches { -- any event
name matches {
DV_TEXT matches {...}
}
data matches {
ITEM_LIST[at0003] matches { -- systemic arterial BP
count matches {|>=2|}
items matches {
ELEMENT[at0004] matches { -- systolic BP
name matches {
DV_TEXT matches {...}
}
value matches {
magnitude matches {...}
}
}
ELEMENT[at0005] matches { -- diastolic BP
name matches {
DV_TEXT matches {...}
}
value matches {
magnitude matches {...}
}
}
}
}
}
}
}
}
}
}The following path extracted from the archetype refers to the systolic blood pressure magnitude:
/data/events[at0006]/data/items[at0004]/value/magnitude
The codes [atnnnn] at each node of the archetype become the archetype_node_id found in each node in the data.
Now consider an OBSERVATION instance (expressed here in ODIN syntax), in which a history of two blood pressures has been recorded using this archetype:
< -- OBSERVATION - blood pressure measurement
archetype_node_id = <"openEHR-EHR-OBSERVATION.blood_pressure.v1">
name = <value = <"BP measurement">>
data = < -- HISTORY
archetype_node_id = <"at0001">
origin = <2005-12-03T09:22:00>
events = < -- List <EVENT>
[1] = < -- EVENT
archetype_node_id = <"at0006">
name = <value = <"sitting">>
time = <2005-12-03T09:22:00>
data = < -- ITEM_LIST
archetype_node_id = <"at0003">
items = < -- List<ELEMENT>
[1] = <
name = <value = <"systolic">>
archetype_node_id = <"at0004">
value = <magnitude = <120.0> ...>
>
[2] = <
name = <value = <"diastolic">>
archetype_node_id = <"at0005">
value = <magnitude = <80.0> ...>
>
>
>
>
[2] = < -- EVENT
archetype_node_id = <"at0006">
name = <value = <"standing">>
time = <2005-12-03T09:27:00>
data = < -- ITEM_LIST
archetype_node_id = <"at0003">
items = < -- List<ELEMENT>
[1] = <
name = <value = <"systolic">>
archetype_node_id = <"at0004">
value = <magnitude = <105.0> ...>
>
[2] = <
name = <value = <"diastolic">>
archetype_node_id = <"at0005">
value = <magnitude = <70.0> ...>
>
>
>
>
>
>
>The same data are shown in JSON syntax:
{
"_type": "OBSERVATION",
"archetype_node_id": "openEHR-EHR-OBSERVATION.blood_pressure.v1",
"name": {
"value": "BP measurement"
},
"data": {
"archetype_node_id": "at0001",
"origin": "2005-12-03T09:22:00",
"events": [
{
"_type": "POINT_EVENT",
"archetype_node_id": "at0006",
"name": {
"value": "sitting"
},
"time": "2005-12-03T09:22:00",
"data": {
"_type": "ITEM_LIST",
"archetype_node_id": "at0003",
"items": [
{
"name": {
"value": "systolic"
},
"archetype_node_id": "at0004",
"value": {
"magnitude": 120.0
}
},
{
"name": {
"value": "diastolic"
},
"archetype_node_id": "at0005",
"value": {
"magnitude": 80.0
}
}
]
}
},
{
"_type": "POINT_EVENT",
"archetype_node_id": "at0006",
"name": {
"value": "standing"
},
"time": "2005-12-03T09:27:00",
"data": {
"_type": "ITEM_LIST",
"archetype_node_id": "at0003",
"items": [
{
"name": {
"value": "systolic"
},
"archetype_node_id": "at0004",
"value": {
"magnitude": 105.0
}
},
{
"name": {
"value": "diastolic"
},
"archetype_node_id": "at0005",
"value": {
"magnitude": 70.0
}
}
]
}
}
]
}
}|
Note
|
in the above example, name values are shown as if they were all DV_TEXTs, whereas in reality in openEHR they more likely to be DV_CODED_TEXT instances; either is allowed by the archetype. This has been done to reduce the size of the example, and makes no difference to the paths shown below.
|
The archetype path mentioned above matches both systolic pressures in the recording. In many querying situations, this may be the intention. However, to uniquely match each of the systolic pressure nodes, paths would need to be created that are based not only on the archetype_node_id but also on another attribute. In the case above, the name attribute may be used, if it is known to have been reliably populated with unique values across sets of immediate siblings under container attributes. The paths are created using the openEHR shortcut form of the `name/value' predicate described earlier, as follows:
/data/events[at0006, 'sitting']/data/items[at0004]/value/magnitude /data/events[at0006, 'sitting']/data/items[at0005]/value/magnitude /data/events[at0006, 'standing']/data/items[at0004]/value/magnitude /data/events[at0006, 'standing']/data/items[at0005]/value/magnitude
Each of these paths has an Xpath equivalent of the following form:
/data/events[@archetype_node_id='at0006' and name/value='standing']/data/items[@archetype_node_id='at0004']/value/magnitude
To achieve unique paths based on the LOCATABLE.name attribute, the system has to specifically ensure uniqueness of name for sibling nodes, e.g. by systematically being set to a copy of one or more other attribute values. For example, in an EVENT object, name could be a string copy of the time attribute.
In general, uniqueness of property values of sibling nodes is not required, and the only guaranteed unique paths are those based on positional predicates.
11.2.4.3. Using Positional Parameters
If it is known within a system that the order of items in container attributes in the data is always preserved across storage, transformation etc, guaranteed unique paths can be created using the Xpath positional parameter. Using the above example, unique to the systolic and diastolic pressures of each event (sitting and standing measurements) can be constructed using the following expressions (identical in openEHR and Xpath):
/data/events[1]/data/items[1]/value/magnitude /data/events[1]/data/items[2]/value/magnitude /data/events[2]/data/items[1]/value/magnitude /data/events[2]/data/items[2]/value/magnitude
11.3. EHR URIs
There are two broad categories of URIs that can be used with any resource: direct references, and queries. The first kind are usually generated by the system containing the referred-to item, and passed to other systems as definitive references, while the second are queries from the requesting system in the form of a URI.
Query-oriented URIs are not formally defined here, since the expectation is that a query service will be used, and that URI formats for querying will dependent on the type of service (for example REST URIs are usually based on served resources).
A dedicated type DV_EHR_URI is defined within the RM data_types package to carry the URIs described here. A DV_EHR_URI instance can only refer to an entity within an openEHR EHR (i.e. not some other kind of resource).
The following guiding principles have been used to inform the design of EHR URIs.
-
It is assumed that one URI 'scheme' (i.e. what precedes the ':' in an IETF RFC3986 URI) is used for each major category of data, i.e. EHR, demographics, etc. Thus, the
ehrscheme corresponds to EHR content. -
URIs described here refer to information items within
VERSION.data, i.e. to objects such asCOMPOSITIONorFOLDER; -
Versions are identified within URIs either via the relevant
VERSIONED_OBJECT.uid(i.e. a GUID) or theVERSION.uid(a 3-partOBJECT_VERSION_ID).
11.3.1. EHR Reference URIs
To create a reference to a node in an EHR in the form of a URI (uniform resource identifier), three elements are needed: the path within a top-level structure, a reference to a top-level structure within an EHR, a reference to an EHR, and an optional reference to an EHR system (i.e. repository). These can be combined to form a URI in an 'ehr' scheme-space which conforms to the following model:
ehr://system_id/ehr_id/top_level_structure_locator/path_inside_top_level_structure
// ----------- variations -----------
ehr://system_id/ehr_id // refer to an EHR within a specific EHR system/service
ehr:/ehr_id // refer to an EHR within the 'current' (i.e. local) EHR system
ehr:/ehr_id/top_level_structure_locator // a specific COMPOSITION, FOLDER etc
ehr:/ehr_id/top_level_structure_locator/path_inside_top_level_structure
// a sub-item of a specific COMPOSITION, FOLDER etcThe possible values for top_level_structure_locator come from attribute names of the class EHR, visible in the https://specifications.openehr.org/releases/RM/latest/ehr.html#ehr_package[ehr package^], namely _compositions, directory etc.
In this way, any object in any openEHR EHR is addressable via a URI. Within ehr space, URL-style references to particular servers, hosts etc are not used, due to not being reliable in the long term. Instead, logical identifiers for EHRs and/or subjects are used, ensuring that URIs remain correct for the lifetime of the resources to which they refer. The openEHR data type DV_EHR_URI is designed to carry URIs of this form, enabling URIs to be constructed for use within LINKs and elsewhere in the openEHR EHR.
|
Note
|
So-called 'plain-text URIs' that contain RFC-3986 forbidden characters such as spaces etc, are allowed on the basis of human readability, but must be RFC-3986 encoded prior to use in e.g. REST APIs or other contexts relying on machine-level conformance. |
See RFC-3986, Universal Resource Identifiers in WWW by Tim Berners-Lee. See W3C document Naming and Addressing: URIs, URLs, … for a starting point on URIs.
An ehr: URI implies the availability of a name resolution mechanism in ehr-space, similar to the DNS, which provides such services for http-, ftp- and other well-known URI schemes. Until such services are established, ad hoc means of dealing with ehr: URIs are likely to be used, as well as more traditional http:// style references. The subsections below describe how URIs of both kinds can be constructed.
11.3.1.1. EHR Location
In ehr-space, a direct locator for an EHR is an EHR identifier (i.e. EHR.ehr_id) as distinct from a subject or patient identifier. Normally the copy in the 'local system' is the one required, and a majority of the time, may be the only one in existence. In this case, the required EHR can be identified simply by an unqualified identifier, giving a URI of the form:
ehr:/347a5490-55ee-4da9-b91a-9bba710f730e/However, due to copying / synchronising of the EHR for one subject among multiple EHR systems, a given EHR identifier may exist at more than one location. It is not guaranteed that each such EHR is a completely identical copy of the others, since partial copying is allowed. Therefore, in an environment where EHR copies exist, and there is a need to identify exactly which EHR instance is required, a system identifier is also required, giving a URI of the form:
ehr://rmh.nhs.net/347a5490-55ee-4da9-b91a-9bba710f730e/11.3.1.2. Top-level Structure Locator
There are two logical ways to identify a top-level structure in an openEHR EHR. The first is via the identifier of the required top-level object (i.e. VERSIONED_OBJECT.uid). When a URI uses the object identifier, the latest trunk version is always assumed. This leads to URIs like the following:
ehr:/347a5490-55ee-4da9-b91a-9bba710f730e/compositions/87284370-2D4B-4e3d-A3F3-F303D2F4F34B
ehr:/347a5490-55ee-4da9-b91a-9bba710f730e/directoryThe second way to identify a top-level structure is by using an exact Version identifier, which takes the form object_id::creating_system_id::version_tree_id. This leads to URIs like the following:
ehr:/347a5490-55ee-4da9-b91a-9bba710f730e/compositions/87284370-2D4B-4e3d-A3F3-F303D2F4F34B::rmh.nhs.net::2This URI identifies a top-level item whose version identifier is 87284370-2D4B-4e3d-A3F3-F303D2F4F34B::rmh.nhs.net::2, i.e. the second trunk version of the Versioned Object indentified by the GUID, created at an EHR system identified by net.nhs.rmh. Note that the mention of a system in the version identifier does not imply that the requested EHR is at that system, only that the top-level object being sought was created at that system.
11.3.1.3. Item URIs
With the addition of path expressions as described earlier, URIs can be constructed that refer to the finest grained items in the openEHR EHR, such as the following:
ehr:/347a5490-55ee-4da9-b91a-9bba710f730e/compositions/87284370-2D4B-4e3d-A3F3-F303D2F4F34B/content[openEHR-EHR-SECTION.vital_signs.v1]/items[openEHR-EHR-OBSERVATION.heart_rate-pulse.v1]/data/events[at0006, 'any event']/data/items[at0004]11.3.1.4. Relative URIs
URIs can also be constructed relative to the current EHR, in which case they do not mention the EHR id, as in the following example:
ehr:compositions/87284370-2D4B-4e3d-A3F3-F303D2F4F34B/content[openEHR-EHR-SECTION.vital_signs.v1]/items[openEHR-EHR-OBSERVATION.blood_pressure.v1]/data/events[at0006, 'any event']/data/items[at0004]
ehr:directory12. Terminology in openEHR
12.1. Overview
openEHR archetypes provide a powerful way to define the meaning of clinical and related data, and to connect, or "bind", data to recognised terminologies such as LOINC, ICDx, ICPC, SNOMED CT and the many other terminologies and vocabularies used in healthcare. Terminology is used in openEHR in the following ways:
-
The values of coded attributes in the reference model are defined by an "openEHR" terminology.
-
Each archetype contains its own internal terminology, defining the meaning of each element.
-
Bindings to external terminologies can be included in an archetype, allowing direct mappings to terms, or mappings to queries that return specific value sets.
-
Querying the EHR using external terminologies is supported by archetype bindings.
The following sections describe these features.
12.2. Terminology to Support the Reference Model
openEHR has its own small terminology and code sets, which are used to provide the value sets of a
number of attributes in the reference model. Code sets are used to express well-known internationally
standardised lists of codes where the codes themselves have meaningful values e.g. the ISO 3166
country codes ("au", "cn", "pl" etc). Six such code sets are used by various attributes in the reference
model, each of type CODE_PHRASE (the openEHR type used to represent a term code).
For other coded attributes, such as PARTICIPATION.function in the reference model, the openEHR
terminology takes the more orthodox route in terminology design, and defines value sets in groups
using meaningless codes and rubrics. These attributes are always of type DV_CODED_TEXT; the code
itself is contained within the defining_code attribute.
The openEHR terminology is described in the openEHR Terminology specification, with computable expressions available at the openEHR terminology Git repository.
12.3. Archetype Internal Terminology
Archetypes contain their own local terminology (found in the 'ontology' section of an archetype). The use of internal term sets is appropriate when there is no structure to the terms (ie no relationships) and when synonyms are not important. Thus, the use is limited to small flat lists of terms. The advantages of the terms being internal to the archetype, apart from computational efficiency mentioned above, are:
-
Queries can be based on archetypes alone and do not require interacting with a terminology server;
-
Translation of the terms is made within an explicit thematic context (since every archetype is about a specific topic) and is therefore far more likely to be accurate;
-
Many terms required in archetypes are not available even in very large terminologies;
-
People can share data based on archetypes even if they do not share terminologies.
It is clear, however, that many archetypes require a connection to external terminologies to provide
the full benefits of automatic processing; this is described in the next section.
The internal terminology takes the form of a set of {code, text, description} semantic definitions for
each node of the archetype structure. Each such term is identified by an "at" (archetype term) code,
e.g. [at0012]. Each code defined locally in an archetype is used for one of two purposes:
-
either to semantically identify the data nodes of the archetype (i.e. to "name" the data), or
-
to provide value-sets for leaf attributes.
For example, the local codes in an "Apgar result" archetype could contain terms for "1 minute
event" and "2 minute event". These codes are associated with the reference model nodes within the
'definition' part of the archetype. In the Apgar example, the two codes (say [at0003] and [at0026])
will be mapped to nodes of reference model type EVENT (rm.data_structures.history package),
as shown below. It is this mapping that is the basis for archetype paths: an archetype
path is simply the alternating pattern of reference model attribute names and node codes.
OBSERVATION[at0000] matches { -- Apgar score
data matches {
HISTORY[at0002] matches { -- history
events cardinality matches {1..*; unordered} matches {
POINT_EVENT[at0003] occurrences matches {0..1} matches {-- 1 minute
offset matches {|PT1M|}
data matches {
ITEM_LIST[at0001] matches {-- structure
items cardinality matches {0..1; ordered} matches {
ELEMENT[at0005] occurrences matches {0..1} matches {-- Heart r
value matches {
ORDINAL matches {
value matches {
0|[local::at0006], -- No heart beat
1|[local::at0007], -- Less than 100 bpm
2|[local::at0008] -- Greater than 100 bpm
}
}
}
}
}
}
}
}
POINT_EVENT[at0026] occurrences matches {0..1} matches {-- 2 minute
offset matches {|PT2M|}
data matches {
use_node ITEM_LIST /data[at0002]/events[at0003]/data[at0001]
...
}
}
}
}
}
}
...The second use of local codes is as values. Above, the ELEMENT node identified by code
[at0005] has as its value constraint an ORDINAL type whose values can be 0, 1, or 2. Each of these
values is coded by the codes [at0006], [at0007], and [at0008]. An extract of the archetype ontology
showing these terms is shown below.
ontology
primary_language = <"en">
languages_available = <"en", "en-us">
terminologies_available = <"LNC205", ...>
term_definitions = <
["en"] = <
items = <
["at0000"] = <
description = <"Clinical score derived from assessment of
breathing, colour, muscle tone, heart rate and reflex
response usually taken at 1, 5 and 10 minutes after birth">
text = <"Apgar score">
>
["at0003"] = <
description = <"Apgar score at one minute">
text = <"1 minute">
>
["at0006"] = <
description = <"No heart beat is present (palpation at base of
umbilical cord)">
text = <"No heart beat">
>
["at0007"] = <
description = <"Heart rate of less than 100 beats per minute">
text = <"Less than 100 beats per minute">
>
["at0008"] = <
description = <"Heart rate of greater than or equal to 100
beats per minute">
text = <"Greater than 100 beats per minute">
>
["at0026"] = <
description = <"Apgar score 2 minutes after birth">
text = <"2 minute">
>
>
>
>
term_bindings = <
["LNC205"] = <
items = <
["/data[at0002]/events[at0003]/data/items[at0025]"] = <[LNC205::9272-6]> -- 1 minute total
["/data[at0002]/events[at0026]/data/items[at0025]"] = <[LNC205::9271-8]> -- 2 minute total
>
>12.4. Binding to External Terminologies
12.4.1. Binding External Terminology Codes to Archetype Codes
The first kind of binding is the ability within an archetype to map an internal code to a code from an
external terminology. The bindings are grouped on the basis of external terminology, allowing any
given internal code in an archetype to be bound to codes in multiple terminologies. Usually, coverage
provided by external terminologies is incomplete, and the mappings may be approximate, so care
must be taken in creating the mappings in the first place. In the example shown above, two
paths are shown respectively as being bound to LOINC codes for 1-minute and 2-minute Apgar total.
In this example, the whole path is bound, meaning that the mapping only holds between [at0025] and
[LNC205::9272-6] when [at0025] occurs in the first path; when it occurs in the second path, the mapping
is to a different LOINC code. This is how so-called "pre-coordinated" codes from external terminologies
can be mapped to an openEHR archetype concept.
Bindings can also be made between atomic internal codes and external codes, in which case the meaning is that the mapping always holds, no matter how many times the internal code is used within the archetype.
12.4.1.1. Binding Terminology Value-sets to Archetypes
An important requirement with respect to terminology is that of specifying value sets for attributes defined in archetypes. Sometimes value sets are defined locally within the archetype, because the terms are not available in published terminologies, and in any case may be too hard to define therein, due to the lack of encapsulation. The terms "no effort", "moderate effort" and "crying" for example are recognised values for the "breathing" attribute of an Apgar result1. In the context of Apgar / breathing, the meanings are clear; clearly however a term with this rubric within a terminology like SNOMED-CT would need to be pre-coordinated. More importantly, there seems to be little business value in mapping a SNOMED term for "no effort", since a query for items containing "no effort" is unlikely to be useful in a clinical context.
For many other kinds of attributes however, terminologies are an appropriate source of values. Often such attributes define kinds of real world phenomena, such as kinds of disease and blood groups, rather than qualities of a phenomenon such as "no effort", or "blue". For these attributes a different kind of connection to external terminology is required. This is achieved in a similar way as for single code bindings: an internal code is defined, in this case an "ac" code ("ac" = archetype constraint), and this is bound to queries to one or more external terminologies, whose result would be a (possibly structured) value set from that terminology. The logical scheme is illustrated in the figure below, where he attribute value to be coded is "blood group phenotype".

Currently there is no standard for such queries. This does not affect archetypes directly, since they simply hold an identifier for a query; the query itself is defined within a "terminology query server". The result of this query is a list of blood group phenotypes, which might appear as shown at the bottom of Figure 40.
12.5. Querying using External Terminologies
Querying through EHR data is frequently cited to be the major utility of terminology with respect to
health information. With the mappings defined in archetypes, a number of approaches are possible,
however the semantics of the intended query need to be understood first. Consider a query for "adenocarcinoma"
on a patient record. SNOMED-CT includes 63 terms beginning with "adenocarcinoma"
(and 171 terms which include the word as a secondary part of the phrase), some as children of a common
parent. Nevertheless, the terms do not all have a single common parent; a choice has to be made
of which terms correspond to the intent of the query. If it is to find any previous diagnosis of "adenocarcinoma",
then at least the terms of the form [snomed-ct::254626006|adenocarcinoma of lung|],
"… of liver" have to be included. These are within the "clinical finding" hierarchy, so the use of these
latter terms should ensure that matches are not made with other uses of the same terms in the record,
e.g. "fear of adenocarcinoma" or "minimal risk of adenocarcinoma". Such correct matching is completely
dependent upon the correct use of SNOMED-CT terms in the first place by the software application
and/or user creating the data. It is easy to imagine an application that saves data (including
openEHR data) in the form of two name/value pairs: <"principal diagnosis", [snomed-ct::35917007|adenocarcinoma|])> and <"site", "lung">. Querying using [snomedct::254626006|adenocarcinoma of lung|] will fail, even though this is exactly the meaning of the data. The data are not wrong
as such, but the lesson is clear: coding of data and code use in queries must be governed by common
models, otherwise there is no hope of reliably processing the data.
Under the openEHR aproach, path-based querying can be used to specify (for example):
-
find
EVALUATIONsbased on aproblem-diagnosis-histological_stagingarchetype with a value at the path/data/items[at0002.1.1]/value/code(histological diagnosis) equal-to-or-subsumed-by "clinical finding" and equal-to or-subsumed-by "adenocarcinoma".
The assumption here is that the value at this path was originally restricted by the archetype from which the path is taken, to conforming to the relation {is-a "clinical finding" and is-a "abnormal morphological mass"}. Any finding of adenocarcinoma of the lung is then forced to be from the resulting subsumption hierarchy; other "adenocarcinoma" terms cannot be wrongly used in this position.
However, even if the archetype had not restricted the value in this way, the same query which searched for any "adenocarcinoma" term at the same path could reasonably be used to locate "previous diagnoses of adenocarcinoma", since this is the only use of the archetype. In a similar way, archetype path-based querying can be used to distinguish the other potential ambiguities described in the section on Section 6.4.
13. Deployment
13.1. 5-tier System Architecture
Previous sections have described the software architecture of the openEHR specifications. Here we describe how the package architecture can be applied to building real systems. The general architectural approach in any openEHR system can be considered as 5 layers (i.e. a "5-tier" architecture). The tiers are as follows.
-
persistence: data storage and retrieval.
-
back-end services: including EHR, demographics, terminology, archetypes, security, record location, and so on. In this layer, the separation of the different services is transparent, and each service has a coarse-grained service interface.
-
virtual EHR: this tier is the middleware, and consists of a coherent set of APIs to the various back-end services providing access to the relevant services, thereby allowing user access to the EHR; including EHR, demographics, security, terminology, and archetype services. It also contains an archetype- and template-enabled kernel, the component responsible for creating and processing archetype-enabled data. In this tier, the separation of backend services is hidden, only the functionality is exposed. Other virtual clients are possible, consisting of APIs for other combinations of back-end services.
-
application logic: this tier consists of whatever logic is specific to an application, which might be a user application, or another service such as a query engine.
-
presentation layer: this layer consists of the graphical interface of the application, where applicable.
The same tiers can be used in large deployments, as shown in the next figure or simply as layers in single- machine applications.
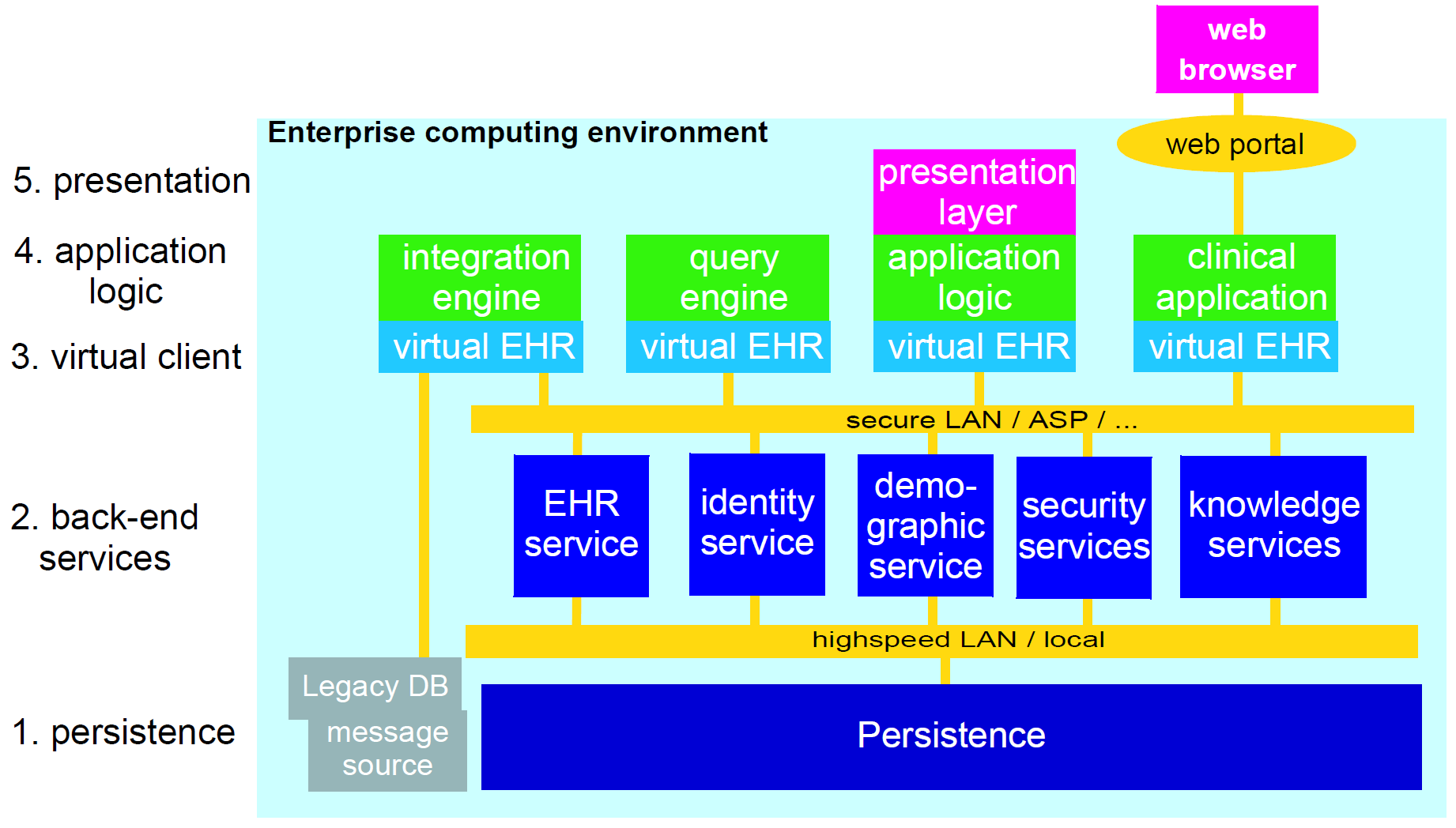
The figure below illustrates an approximate mapping of major parts of the openEHR software architecture to the 5-tier scheme. Clearly where parts of the architecture are used will depend on various implementation choices; the mapping shown is therefore not definitive. Nevertheless, the principal use of parts of the architecture is likely to be similar in most systems, as follows:
-
RM and AM: mainly used to construct an archetype- and template-processing kernel;
-
RM
common.change_controlpackage: provides the logic for versioning in versioned services such as the EHR and demographics; -
SM: various service model packages define the exposed interfaces of major services;
-
SM
virtual_ehrpackage defines the API of the virtual EHR component; -
archetypes: archetypes might be assumed directly in some applications, e.g. a specialist peri-natal package might be partly based on a family of archetypes for this specialisation;
-
templates: both archetypes and templates will be used in the presentation layer of applications.
Some will base the GUI code on them, while others will have either tool-generated code, or dynamically generate forms based on particular templates and archetypes. In the future, an abstract persistence API and optimised persistence models (transformations of the existing RM models) are likely to be published by openEHR in order to help with the implementation of databases.
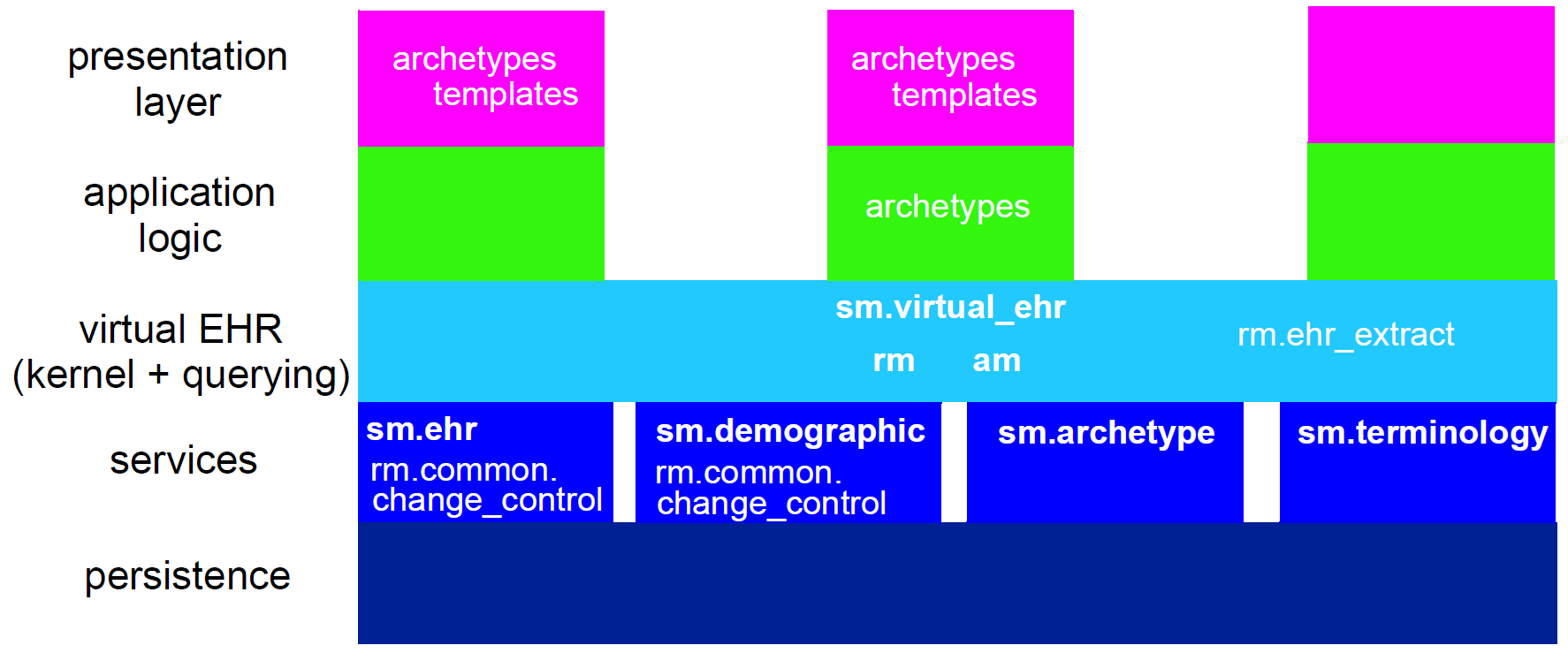
14. Integrating openEHR with other Systems
14.1. Overview
Getting data in and out of the EHR is one of the most basic requirements openEHR aims to satisfy. In "greenfield" (new build) situations, and for data being created by GUI applications via the openEHR EHR APIs, there is no issue, since native openEHR structures and semantics are being used. In almost all other situations, existing data sources and sinks have to be accounted for. In general, external or ‘legacy’ data (here the term is used for convenience, and does not imply anything about the age or quality of the systems in question) have different syntactic and semantic formats than openEHR data, and seamless conversion requires addressing both levels.
Existing data sources and sinks include relational databases, HL7v2 messages, HL7 CDA documents and are likely to include ISO 13606 data. HL7v2 messages are probably one of the most common sources of pathology messages in many countries; EDIFACT messages are another. More recently, HL7v2 messages have been designed for referrals and even discharge summaries. Not all legacy systems are standardised; most hospital and GP products have their own private models of data and terminology usage.
The primary need with respect to legacy data is to be able to convert data from multiple mutually incompatible sources into a single, standardised patient-centric EHR for each patient, that can then be longitudinally viewed and queried. This is what enables GP and specialist notes, diagnoses and plans to be integrated with laboratory results from multiple sources, patient notes, administrative data and so on, to provide a coherent record of the patient journey.
In technical terms, a number of types of incompatibility have to be dealt with. There is no guarantee
of correspondence of scope of incoming transactions and target openEHR structures - an incoming
document for example might correspond to a number of clinical archetypes. Structure will not usually
correspond, with legacy data (particularly messages) usually having flatter structures than those
defined in target archetypes. Terminology use is extremely variable in existing systems and messages,
and also has to be dealt with. Data types will also not correspond directly, so that for example, a mapping
between an incoming string "110/80 mmHg" and the target openEHR form of two
DV_QUANTITY objects each with their own value and units has to be made.
14.2. Integration Archetypes
The foundation of a key approach to the integration problem is the use of two kinds of archetypes. So far in this document "archetypes" has meant "designed" archetypes, generally clinical, demographic or administrative. The common factors for all such archetypes are:
-
they are based on the main part of the reference model, particularly the Entry subtypes
OBSERVATION,EVALUATION,INSTRUCTIONandACTION; -
they are consciously designed from scratch by groups of domain specialists, and integrated into the existing library of openEHR archetypes;
-
there is one archetype per identifiable health "concept", such as an observation type, person type etc.
A second category of archetypes is "integration" archetypes. These are characterised as follows:
-
they are based on the same high-level types (
COMPOSITION,SECTIONetc), but use the Entry subtypeGENERIC_ENTRY(see EHR Information Model); -
they are designed to mimic the structure of legacy or existing data or messages; the design effort therefore is completely different, and is more likely to be done by IT or other technical staff who are familiar with the structures of the incoming data;
-
there is one integration archetype per message type or identifiable source data that makes sense as a transaction to the EHR.
In the data integration environment, "designed" archetypes always define the target structures, coding and other semantics of data, while "integration" archetypes provide the means mapping of external data into the openEHR environment.
14.3. Data Conversion Architecture
The integration archetype-based strategy for importing data into an openEHR system, shown below, consists of two steps.
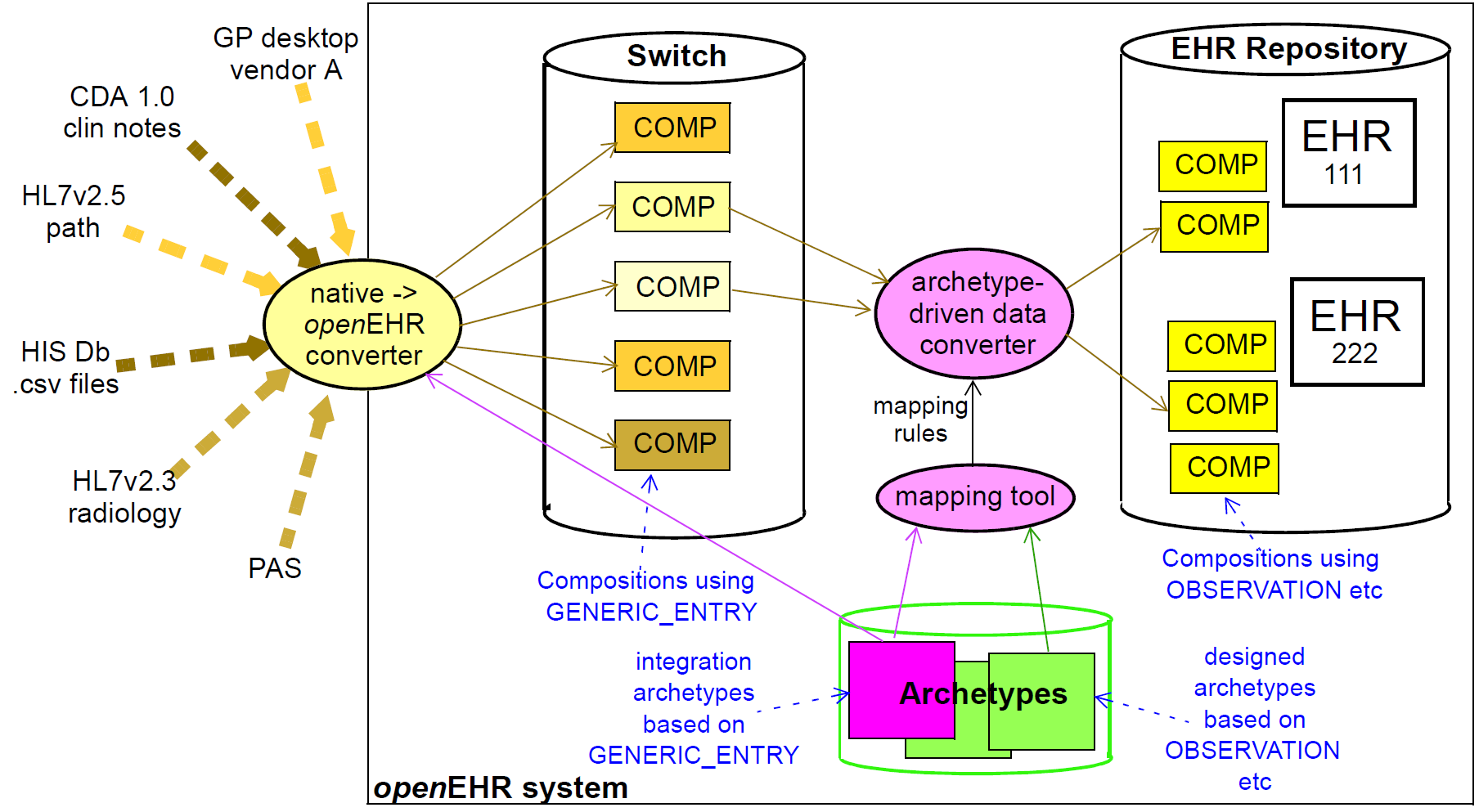
Firstly, data are converted from their original syntactic format into openEHR COMPOSITION / SECTION / GENERIC_ENTRY structures, shown in the openEHR integration switch. Most of the data will
appear in the GENERIC_ENTRY part, controlled by an integration archetype designed to mimic the
incoming structure (such as an HL7v2 lab message) as closely as possible; FEEDER_AUDIT structures
are used to contain integration meta-data. The result of this step is data that are expressed in the
openEHR type system (i.e. as instances of the openEHR reference model), and are immediately amenable
to processing with normal openEHR software.
In the second step, semantic transformation is effected, by the use of mappings between integration and designed archetypes. Such mappings are created by archetype authors using tools. The mapping rules are the key to defining structural transformations, use of terminological codes, and other changes. Serious challenges of course remain in the business of integrating heterogeneous systems; some of these are dealt with in the Common IM document sections on Feeder systems.
15. Relationship to Standards
The openEHR specifications make use of available standards where relevant, and as far as possible in a compatible way. However, for the many standards have never been validated in their published form (i.e. the form published is not tested in implementations, and may contain errors), openEHR makes adjustments so as to ensure quality and coherence of the openEHR models. In general, "using" a standard in openEHR may mean defining a set of classes which map it into the openEHR type system, or wrap it or express it in some other compatible way, allowing developers to build completely coherent openEHR systems, while retaining compliance or compatibility with standards. The standards relevant to openEHR fall into a number of categories as follows.
15.1. Standards by which openEHR can be evaluated
These standards define high-level requirements or compliance criteria which can be used to provide a means of normative comparison of openEHR with other related specifications or systems:
15.2. Standards which have influenced the design of openEHR specifications
The following standards have influenced the design of the openEHR specifications:
-
OMG HDTF Standards - general design
-
CEN HISA 12967-3: Health Informatics Service Architecture - Computational viewpoint
15.3. Standards which have influenced the design of openEHR archetypes
The following standards are mainly domain-level models of clinical practice or concepts, and are being used to design openEHR archetypes and templates.
-
CEN HISA 12967-2: Health Informatics Service Architecture - Information viewpoint
-
CEN ENV 13940: Continuity of Care.
15.4. Standards which are used "inside" openEHR
The following standards are used or referenced at a fine-grained level in openEHR:
-
ISO 8601: Syntax for expressing dates and times (used in openEHR Quantity package)
-
Unified Coding for Units of Measure (UCUM) (used by openEHR Quantity data type)
-
HL7v3 GTS: General Timing Specification syntax (used by openEHR Time specification data types).
-
some HL7v3 domain vocabularies are mapped to the openEHR terminology.
-
IETF RFC4880 - openPGP.
15.5. Standards which require a conversion gateway
The following standards are in use and require data conversion for use with openEHR:
-
ISO 13606:2005 - Electronic Health Record Communication - near-direct conversion possible, as openEHR and ISO 13606 are actively maintained to be compatible.
-
HL7v3 CDA: Clinical Document Architecture (CDA) release 2.0 - fairly close conversion may be possible.
-
HL7v3 messages. Quality of conversion currently unknown due to flux in HL7v3 messaging specifications and diversity of message schemas.
-
HL7v2 messages. Importing of HL7v2 message data is technically not difficult, and is already used in some openEHR systems. Export from openEHR may also be possible.
16. Implementation Technology Specifications
16.1. Overview
ITSs are created by the application of transformation rules from the "full-strength" semantics of the abstract models to equivalents in a particular technology. Transformation rules usually include mappings of:
-
names of classes and attributes;
-
property and function signature mapping;
-
mapping of basic types e.g. strings, numerics;
-
how to handle multiple inheritance;
-
how to handle generic (template) types;
-
how to handle covariant and contravariant redefinition semantics;
-
the choice of mapping properties with signature
xxxx:T(i.e. properties with no arguments) to stored attributes (xxxx:T) or functions (xxxx():T); -
how to express pre-conditions, post-conditions and class invariants;
-
mappings between assumed types such as List<>, Set<> and inbuilt types.
ITSs are being developed for a number of major implementation technologies, as summarised below. Implementors should always look for an ITS for the technology in question before proceeding. If none exists, it will need to be defined. A methodology to do this is being developed. The figure below illustrates the implementation technology specification space. Each specification documents the mapping from the standard object-oriented semantics used in the openEHR abstract models, and also provides an expression of each of the abstract models in the ITS formalism.
References
Anderson, D. R., & Anderson, J. (1996). Security in Clinical Information Systems. Retrieved from http://www.cl.cam.ac.uk/users/rja14/policy11/policy11.html
Beale, T. (2002). Archetypes: Constraint-based Domain Models for Future-proof Information Systems. In K. Baclawski & H. Kilov (Eds.), Eleventh OOPSLA Workshop on Behavioral Semantics: Serving the Customer (pp. 16–32). Northeastern University, Boston. Retrieved from https://www.openehr.org/publications/archetypes/archetypes_beale_oopsla_2002.pdf
Beale, T., & Heard, S. (2007). An Ontology-based Model of Clinical Information. In K. K. et al. (Ed.), Proceedings MedInfo 2007 (pp. 760–764). IOS Publishing. Retrieved from http://www.openehr.org/publications/health_ict/MedInfo2007-BealeHeard.pdf
Elstein, A. S., Shulman, L. S., & Sprafka, S. A. (1978). Medical problem solving: an analysis of clinical reasoning. Cambridge, MA: Harvard University Press.
Kifer, M., Lausen, G., & Wu, J. (2000). Logical Foundations of Object-Oriented and Frame-Based Languages. Journal of the ACM, 42, 741–843. Retrieved from ftp://ftp.cs.sunysb.edu/pub/TechReports/kifer/flogic.pdf
Maier, M. (2000). Architecting Principles for Systems-of-Systems. University of Alabama in Huntsville. Retrieved from http://www.infoed.com/Open/PAPERS/systems.htm
